VISTA-3D
Important
NVIDIA NIM currently is in limited availability, sign up here to get notified when the latest NIMs are available to download.
The Versatile Imaging SegmenTation and Annotation (VISTA) model combines semantic segmentation with interactivity, offering high accuracy and adaptability across diverse anatomical areas for medical imaging.
VISTA-3D is a domain-specialized interactive foundation model developed for segmenting and annotating human anatomies with precision and has three (3) core workflows:
Segment everything: Enables whole body exploration, crucial for understanding complex diseases affecting multiple organs and for holistic treatment planning.
Segment using class: Provides detailed sectional views based on specific classes, essential for targeted disease analysis or organ mapping, such as tumor identification in critical organs.
Segment point prompts: Enhances segmentation precision through user-directed, click-based selection. This interactive approach accelerates the creation of accurate ground-truth data, essential in medical imaging analysis.
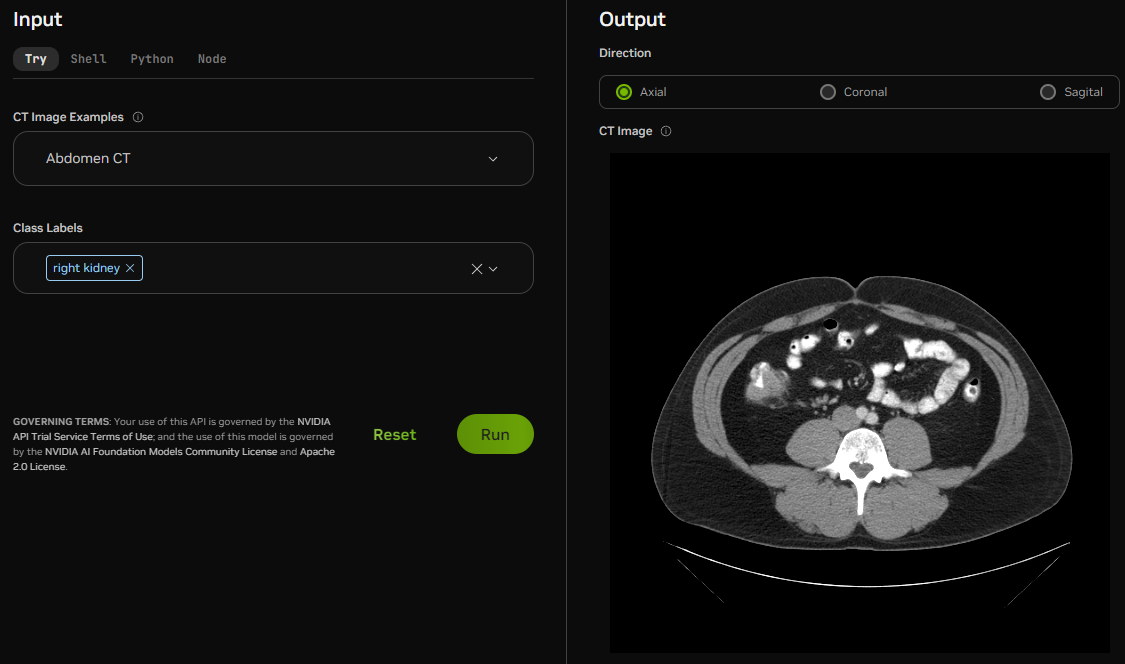
Example output
VISTA-3D is powered by NVIDIA NIM, part of NVIDIA AI Enterprise. This integration offers a suite of easy-to-use microservices designed to accelerate the deployment of AI models, like VISTA-3D, across the cloud, data centers, and workstations. For more information, refer to MONAI.
Note
A more detailed description of the model can be found in the Model Card.
Model Specific Requirements
The following are specific requirements for VISTA-3D NIM.
Important
Please refer to NVIDIA NIM documentation for necessary hardware, operating system, and software prerequisites if you have not done so already
Hardware
Software
Minimum NVIDIA Driver Version: 450.36.06
Quickstart Guide
Note
This page assumes Prerequisite Software (Docker, NGC CLI, NGC registry access) is installed and set up.
Pull the NIM container.
docker pull nvcr.io/nvidia/nim/medical_imaging_vista3d:24.03
Note
The VISTA-3D model is shipped inside of the docker image, therefore no external model download is necessary.
Run the NIM container.
1docker run --rm -it --name vista3d \ 2 --runtime=nvidia -e CUDA_VISIBLE_DEVICES=0 \ 3 --shm-size=8G \ 4 -p 8008:8008 \ 5 nvcr.io/nvidia/nim/medical_imaging_vista3d:24.03
Wait until the gRPC health check returns
truebefore proceeding.curl localhost:8008/health/ready
Run Inference in a new terminal.
1curl -X POST http://0.0.0.0:8008/vista3d/inference \ 2-H "Content-Type: application/json" \ 3--output output.nrrd \ 4-d '{ 5"image": "https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz", 6"prompts": { 7 "classes": ["liver", "spleen"] 8} 9}'
Detailed Instructions
This section provides additional details outside of the scope of the Quickstart guide.
Pull Image
Container image tags can be retrieved using the following command.
ngc registry image info nvcr.io/nvidia/nim/medical_imaging_vista3d
1Image Repository Information 2 Name: medical_imaging_vista3d 3 Display Name: VISTA-3D NIM 4 Short Description: The VISTA-3D NIM Container accelerates ground truth data creation by streamlining segmentation and annotations with an interactive foundation model. 5 Built By: 6 Publisher: NVIDIA 7 Multinode Support: False 8 Multi-Arch Support: False 9 Logo: https://catalog.ngc.nvidia.com/demos/vista.jpg 10 Labels: NVIDIA NIM, NVIDIA AI Enterprise Supported 11 Public: No 12 Access Type: 13 Associated Products: [] 14 Last Updated: Mar 15, 2024 15 Latest Image Size: 8.77 GB 16 Signed Tag?: False 17 Latest Tag: 24.03 18 Tags: 19 24.03
Pull the container image.
docker pull nvcr.io/nvidia/nim/medical_imaging_vista3d:24.03
ngc registry image pull nvcr.io/nvidia/nim/medical_imaging_vista3d:24.03
Launch Microservice
Launch the NVIDIA VISTA-3D container in detached mode. Service will start in a few seconds.
1docker run --rm -it --name vista3d \ 2--runtime=nvidia -e CUDA_VISIBLE_DEVICES=0 \ 3--shm-size=8G \ 4-p 8008:8008 \ 5-d nvcr.io/nvidia/nim/medical_imaging_vista3d:24.03
To observe the progress, please use the docker logs as follows.
docker logs --follow vista3d
The following logs indicate the Nvidia VISTA-3D server is running.
1INFO: Started server process [88] 2INFO: Waiting for application startup. 3INFO: Application startup complete. 4INFO: Uvicorn running on http://0.0.0.0:8008 (Press CTRL+C to quit)
Health and Liveness checks
The container exposes health and liveness endpoints for integration into existing systems such as Kubernetes. These endpoints only return an HTTP 200 OK status code if the service is ready or live, respectively.
Run these in a new terminal.
Readiness
1curl localhost:8008/health/ready
2...
3true
Liveness
1curl localhost:8008/health/live
2...
3true
Run Inference
Inference run on single NIfTI images. You can download any NIfTI data from http://medicaldecathlon.com/ or use a sample image as https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz directly in the inference call or use MONAI core or MONAI label APIs to download data (see MONAI tutorials / APIs).
Download and install 3D Slicer as the Viewer.
The model can be run using four different ways:
Run Vista-3D Model for All Labels
1curl -X POST 'http://0.0.0.0:8008/vista3d/inference' \
2-H 'Content-Type: application/json' \
3--output output1.nrrd \
4-d '{
5"image": "https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz"
6}'
You can verify the output file output1.nrrd along with the input image using 3D Slicer and all organ masks should be present.
Run Vista-3D Model for Certain Organs
1curl -X POST http://0.0.0.0:8008/vista3d/inference \
2-H "Content-Type: application/json" \
3--output output2.nrrd \
4-d '{
5"image": "https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz",
6"prompts": {
7 "classes": ["liver", "spleen"]
8}
9}'
You can verify the output file output2.nrrd along with the input image using 3D Slicer and mask of liver and spleen should be present.
Run Vista-3D Model for the Specific Output Format
1curl -X POST http://0.0.0.0:8008/vista3d/inference \
2-H "Content-Type: application/json" \
3--output output3.nii.gz \
4-d '{
5"image": "https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz",
6"prompts": {
7"classes": ["liver", "spleen"]
8},
9"output": {"extension": ".nii.gz", "dtype": "uint16"}
10}'
You can verify the output file output3.nii.gz along with the input image using 3D Slicer and mask of liver and spleen should be present.
Run the Vista Model for a Single Organ and Provide Guidance With User Points
1curl -X POST http://0.0.0.0:8008/vista3d/inference \
2-H "Content-Type: application/json" \
3--output output4.nrrd \
4-d '{
5"image": "https://assets.ngc.nvidia.com/products/api-catalog/vista3d/example-1.nii.gz",
6"prompts": {
7"points": {"spleen": [[52,181,126], [137,152,171]]}
8}
9}'
You can verify the output file output4.nrrd along with the input image using 3D Slicer and the mask of the spleen should be present.
Stopping the Container
When you’re done testing the endpoint, you can bring down the container by running docker stop vista3d in a new terminal.