Deploy AlphaFold2 NIM
Learn how to run the AlphaFold2 NIM on DGX Cloud Lepton.
AlphaFold2 is a protein structure prediction model from Google DeepMind. AlphaFold2 demonstrates state-of-the-art performance at predicting protein structure from amino acid sequences.
The NVIDIA AlphaFold2 NIM can:
- Predict protein structure given a protein sequence.
- Predict a multiple sequence alignment (MSA) for a given sequence against a series of protein sequence databases. The NIM includes the GPU-accelerated MMSeqs2 package for Multiple Sequence Alignment.
- Predict a protein structure given a pre-computed MSA of the sequence against protein sequence databases.
For more information about AlphaFold2, see the AlphaFold2 paper in Nature. If you use this NIM or AlphaFold2, make sure to cite the paper:
Jumper, J., Evans, R., Pritzel, A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). https://doi.org/10.1038/s41586-021-03819-2
Refer to NVIDIA NIM for AlphaFold2 documentation for more information.
Prerequisites
To run the AlphaFold2 NIM, you need:
- Access to your NIM Container Registry and NGC API Key. For more information, refer to the setup information.
- Access to at least 1x NVIDIA A100 GPU or higher on DGX Cloud Lepton
- At least 1.3 TB of available SSD storage
- At least 24 CPU cores
Create Endpoint from NVIDIA NIM
Navigate to the create endpoint page on the dashboard.
In the NIM Configuration section, specify the container image, registry auth key, and NGC API key.
- Image: Choose Custom image and enter
nim/deepmind/alphafold2:2.1.0. - Registry auth: Select one of the existing registry auth configurations you created in your workspace or create a new one.
- NGC API key: Select one of the existing NGC API keys you saved in your workspace as a secret or create a new one.
The AlphaFold2 NIM can take multiple hours to download the dataset initially. To prevent the endpoint from failing before the model is downloaded, go to Advanced Configuration and set Healthcheck Initial Delay Seconds to Custom with a value of 7200 seconds.
Saving AlphaFold2 Model in Cache
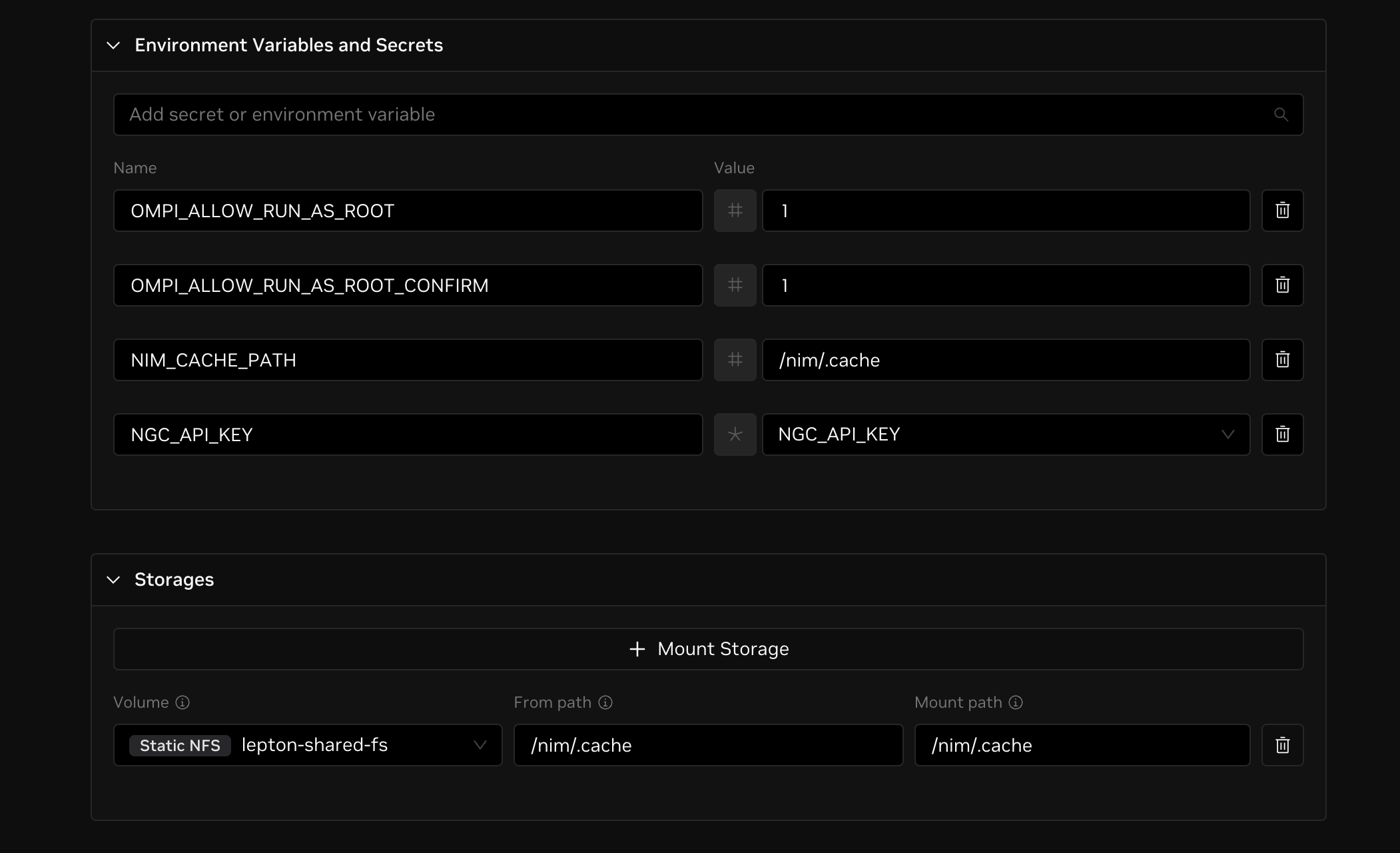
Since the AlphaFold2 model can take hours to download, you may want to store the model in a persistent cache. To do this, set an environment variable with a new cache path in persistent storage and mount that storage path.
Go to Environment Variables and Secrets and click Add Variable. Set the variable name to NIM_CACHE_PATH and the variable value to /nim/.cache. Then go to Storage and click + Mount Storage. Select your desired volume, set the from path to /nim/.cache, and set the to path to /nim/.cache.
This should now look like the image below.
For other endpoint configurations, refer to the Endpoints documentation.
Testing AlphaFold2 Endpoint
Once your endpoint is ready, you can use the playground to send requests directly from the UI, or you can send requests to the endpoint URL by running a command like the following:
If you set your endpoint to be publicly accessible, you do not need to set an endpoint token.