Quickstart Guide for NVIDIA Earth-2 Correction Diffusion NIM#
Use this documentation to get started with NVIDIA Earth-2 Correction Diffusion (CorrDiff) NIM.
Important
Before you can use this documentation, you must satisfy all prerequisites.
Warning
This quick start guide is designed for use with Nvidia’s A100 or H100 GPUs.
GPUs with lower VRAM and/or compute may require EARTH2NIM_TARGET_BATCHSIZE to be lowered or the time out in the post request to be increased.
See the configuration page for different deployment options.
Launching the NIM#
Pull the NIM container with the following command.
Note
The container is ~26GB (uncompressed) and the download time depends on internet connection speeds.
docker pull nvcr.io/nim/nvidia/corrdiff:1.0.0
Run the NIM container with the following command. This command starts the NIM container and exposes port 8000 for the user to interact with the NIM. It pulls the model on the local filesystem.
Note
The model is ~4GB and the download time depends on internet connection speeds.
export NGC_API_KEY=<NGC API Key>
docker run --rm --runtime=nvidia --gpus all --shm-size 2g \
-p 8000:8000 \
-e NGC_API_KEY \
-t nvcr.io/nim/nvidia/corrdiff:1.0.0
After the NIM is running, you see output similar to the following:
I0829 05:18:42.152983 108 grpc_server.cc:2466] "Started GRPCInferenceService at 0.0.0.0:8001"
I0829 05:18:42.153277 108 http_server.cc:4638] "Started HTTPService at 0.0.0.0:8090"
I0829 05:18:42.222628 108 http_server.cc:320] "Started Metrics Service at 0.0.0.0:8002"
Checking NIM Health#
Open a new terminal, leaving the current terminal open with the launched service.
Wait until the health check end point returns
{"status":"ready"}before proceeding. This might take a couple of minutes. Use the following methods to query the health check.
Bash#
curl -X 'GET' \
'http://localhost:8000/v1/health/ready' \
-H 'accept: application/json'
Python#
import requests
r = requests.get("http://localhost:8000/v1/health/ready")
if r.status_code == 200:
print("NIM is healthy!")
else:
print("NIM is not ready!")
Fetching Input Data#
The CorrDiff NIM launches with a default model profile trained for downscaling over the contiguous United States. The input to this model is GEFS weather data at 0.25-degree (~25km) resolution over the United States, sourced from NOAA. CorrDiff use this input data to generate downscaled weather fields at a 3km resolution, similar to NOAA’s HRRR model. For additional details, refer to the model card.
To simplify data preparation, we use Earth2Studio to fetch and format the input data as follows:
Fetch a set of select GEFS variable at a resolution 0.25-degrees on a lat-lon grid.
Fetch a set of regular GEFS variables at a resolution of 0.5-degrees and interpolate to 0.25 degree grid on a lat-lon grid.
Crop both variables to a bounded box over the United States.
Concatenate both sets of variables as well as an integer field that denotes the forecast lead time the input represents.
Use the script below to download and structure the data:
from datetime import datetime, timedelta
import numpy as np
import torch
from earth2studio.data import GEFS_FX, HRRR, GEFS_FX_721x1440
GEFS_SELECT_VARIABLES = [
"u10m",
"v10m",
"t2m",
"r2m",
"sp",
"msl",
"tcwv",
]
GEFS_VARIABLES = [
"u1000",
"u925",
"u850",
"u700",
"u500",
"u250",
"v1000",
"v925",
"v850",
"v700",
"v500",
"v250",
"z1000",
"z925",
"z850",
"z700",
"z500",
"z200",
"t1000",
"t925",
"t850",
"t700",
"t500",
"t100",
"r1000",
"r925",
"r850",
"r700",
"r500",
"r100",
]
ds_gefs = GEFS_FX(cache=True)
ds_gefs_select = GEFS_FX_721x1440(cache=True, product="gec00")
def fetch_input_gefs(
time: datetime, lead_time: timedelta, content_dtype: str = "float32"
):
"""Fetch input GEFS data and place into a single numpy array
Parameters
----------
time : datetime
Time stamp to fetch
lead_time : timedelta
Lead time to fetch
filename : str
File name to save input array to
content_dtype : str
Numpy dtype to save numpy
"""
dtype = np.dtype(getattr(np, content_dtype))
# Fetch high-res select GEFS input data
select_data = ds_gefs_select(time, lead_time, GEFS_SELECT_VARIABLES)
select_data = select_data.values
# Crop to bounding box [225, 21, 300, 53]
select_data = select_data[:, 0, :, 148:277, 900:1201].astype(dtype)
assert select_data.shape == (1, len(GEFS_SELECT_VARIABLES), 129, 301)
# Fetch GEFS input data
pressure_data = ds_gefs(time, lead_time, GEFS_VARIABLES)
# Interpolate to 0.25 grid
pressure_data = torch.nn.functional.interpolate(
torch.Tensor(pressure_data.values),
(len(GEFS_VARIABLES), 721, 1440),
mode="nearest",
)
pressure_data = pressure_data.numpy()
# Crop to bounding box [225, 21, 300, 53]
pressure_data = pressure_data[:, 0, :, 148:277, 900:1201].astype(dtype)
assert pressure_data.shape == (1, len(GEFS_VARIABLES), 129, 301)
# Create lead time field
lead_hour = int(lead_time.total_seconds() // (3 * 60 * 60)) * np.ones(
(1, 1, 129, 301)
).astype(dtype)
input_data = np.concatenate([select_data, pressure_data, lead_hour], axis=1)[None]
return input_data
input_array = fetch_input_gefs(datetime(2023, 1, 1), timedelta(hours=0))
np.save("corrdiff_inputs.npy", input_array)
Running the script will generate the NumPy array corrdiff_inputs.npy, which is now ready to use with the model.
Inference Request#
With the pre-processed input for this CorrDiff US model, the API to the NIM can now be used as follows. The NumPy array is sent to the NIM as a file along with several other parameters that can be used to control the underlying diffusion model. For complete documentation on the API specification of the NIM, visit the API documentation page.
Bash#
curl -X POST \
-F "input_array=@corrdiff_inputs.npy" \
-F "samples=2" \
-F "steps=12" \
-o output.tar \
http://localhost:8000/v1/infer
Python#
import requests
url = "http://localhost:8000/v1/infer"
files = {
"input_array": ("input_array", open("corrdiff_inputs.npy", "rb")),
}
data = {
"samples": 2,
"steps": 14,
"seed": 0,
}
headers = {
"accept": "application/x-tar",
}
print("Sending post request to NIM")
# Adjust time out (seconds) below
r = requests.post(url, headers=headers, data=data, files=files, timeout=180)
if r.status_code != 200:
raise Exception(r.content)
else:
# Dump response to file
with open("output.tar", "wb") as tar:
tar.write(r.content)
Results#
The results are inside a tar file that can be explored using:
tar -tvf output.tar
-rw-r--r-- 0/0 60555392 1970-01-01 00:00 000_000.npy
-rw-r--r-- 0/0 60555392 1970-01-01 00:00 001_000.npy
Tip
The tar archive is populated with NumPy arrays that have the naming convention {sample index}_{batch index}.npy.
Important
For most inference calls, it is important to specify a longer timeout period for a request to this NIM. Refer to the performance data for estimated inference speeds on different GPU models.
Streaming Responses#
The CorrDiff NIM can be used to stream back samples as they are generated in instances where the client requests a larger sample size than the NIMs inference batch size. This can be extremely useful when creating a pipeline that runs following processes on each time-step. The follow snippet can be used to access each time-step, in memory, from the NIM as it is generated.
Note
The environment variable EARTH2NIM_TARGET_BATCHSIZE governs the batch size used inside the NIM. When this value is lower than the requested sample size, streaming the results back can be useful. See the configuration guide for more details on this parameter.
import io
import tarfile
from pathlib import Path
import numpy as np
import requests
import tqdm
url = f"http://localhost:8000/v1/infer"
files = {
"input_array": ("input_array", open("corrdiff_inputs.npy", "rb")),
}
data = {
"samples": 16,
"steps": 8,
"seed": 0,
}
headers = {
"accept": "application/x-tar",
}
pbar = tqdm.tqdm(range(data["samples"]), desc="CorrDiff samples")
with (
requests.post(
url,
headers=headers,
files=files,
data=data,
timeout=300,
stream=True,
)
) as resp:
resp.raise_for_status()
with tarfile.open(fileobj=resp.raw, mode="r|") as tar_stream:
# Loop over file members from tar stream
for member in tar_stream:
arr_file = io.BytesIO()
arr_file.write(tar_stream.extractfile(member).read())
arr_file.seek(0)
data = np.load(arr_file)
# Array names are in the form <BATCH_IDX>_<SAMPLE_IDX>.npy
arr_sample_idx, _ = (
int(x) for x in Path(member.name).stem.split("_", maxsplit=1)
)
pbar.write(f"Received data for sample {arr_sample_idx}")
pbar.write(f"Output numpy {member.name} with shape {data.shape}")
pbar.update(1)
Post Processing#
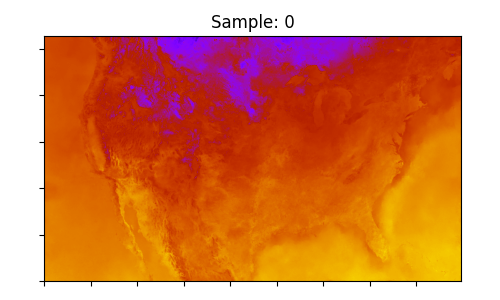
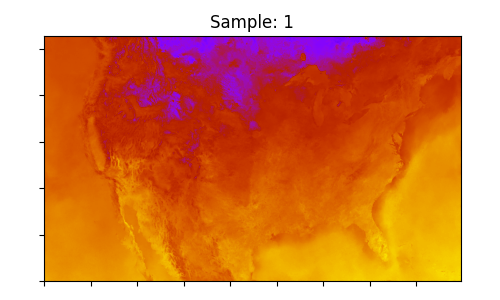
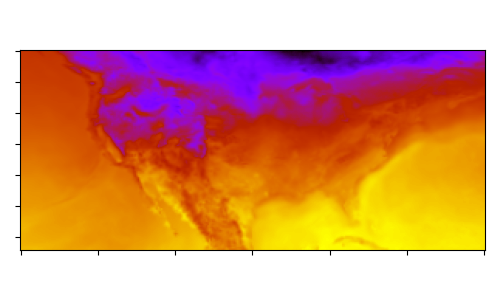
The final step is to perform some basic visualization of the results. The surface temperature is plotted from both the input and output arrays.
Note
The CorrDiff NIM returns the raw output data array. Additional metadata must be processed client side using information in the model card. The latitude and longitude coordinate values can be obtained from the corrdiff_output_lat.npy and corrdiff_output_lon.npy files in the model package.
import matplotlib.pyplot as plt
import numpy as np
import tarfile
import io
# Output variables: [u10m, v10m, t2m, tp, csnow, cicep, cfrzr, crain]
variable = 2
input = np.load("corrdiff_inputs.npy")
fig, ax = plt.subplots(1,1, figsize=(5,3))
ax.imshow(input[0,0,variable], cmap="gnuplot")
ax.set_yticklabels([])
ax.set_xticklabels([])
fig.tight_layout()
plt.savefig("input_t2m.png")
with tarfile.open("output.tar") as tar:
for i, member in enumerate(tar.getmembers()):
arr_file = io.BytesIO()
arr_file.write(tar.extractfile(member).read())
arr_file.seek(0)
data = np.load(arr_file)
plt.close("all")
fig, ax = plt.subplots(1,1, figsize=(5,3))
ax.set_title(f"Sample: {i}")
ax.imshow(data[0,0,variable], origin='lower', cmap="gnuplot")
ax.set_yticklabels([])
ax.set_xticklabels([])
fig.tight_layout()
plt.savefig(f"output_t2m_sample_{i}.png")
Input surface temperature contour at ~25km resolution:
Output surface temperature samples downscaled using CorrDiff to 3km resolution: