Performing Inference with gRPC
Contents
Performing Inference with gRPC#
This section will use the pre-trained BioNeMo checkpoints to demonstrate how to setup a lightweight inference server with gRPC.
Prerequisites#
Linux OS
Pascal, Volta, Turing, or an NVIDIA Ampere architecture-based GPU.
NVIDIA Driver
Docker
Import#
Components for performing inference are part of the BioNeMo source code. This example demonstrates the use of these components.
Prior to execution of this code you will need to connect to the gRPC client. A provided script is included in the framework.
Run the following command to connect for MegaMolBART:
python3 -m bionemo.model.molecule.megamolbart.grpc.service
Similarly, for ESM1 or ProtT5:
python3 -m bionemo.model.protein.esm1nv.grpc.service
python3 -m bionemo.model.protein.prott5nv.grpc.service
Detailed Example with MegaMolBART#
Expanded from bionemo/examples/molecule/megamolbart/nbs/Inference.ipynb
The MegaMolBART inference wrapper implements following functions:
smis_to_hiddensmis_to_embeddinghidden_to_smis
In this generative task, which uses both the encoder and the decoder from the pre-trained model, the embeddings for the input query SMILES will be obtained. Once the embeddings are obtained, they will be used to generate analogs/related designs of small molecules for chemical space exploration.
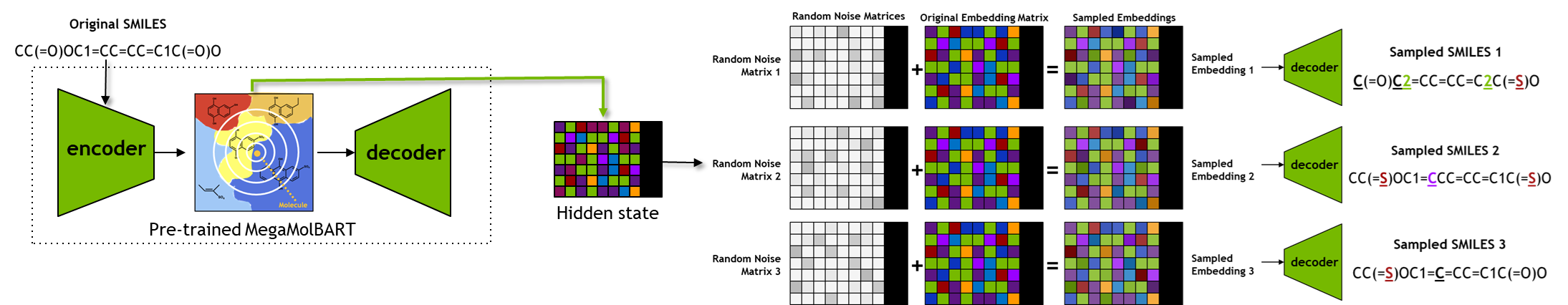
Note
gRPC limits the request size to 4MB.
import logging
import warnings
warnings.filterwarnings('ignore')
warnings.simplefilter('ignore')
Setup and Test Data#
The InferenceWrapper is an adaptor that allows interaction with inference service. The script for creating this class has been included in detail here for the MegaMolBART example at bionemo.examples.molecule.megamolbart.nbs.infer .
import grpc
import torch
import logging
from megamolbart_pb2_grpc import GenerativeSamplerStub
from megamolbart_pb2 import InputSpec
log = logging.getLogger(__name__)
class InferenceWrapper():
def __init__(self):
channel = grpc.insecure_channel('localhost:50051')
self.stub = GenerativeSamplerStub(channel)
def smis_to_embedding(self, smis):
spec = InputSpec(smis=smis)
resp = self.stub.SmilesToEmbedding(spec)
embeddings = torch.FloatTensor(list(resp.embeddings))
embeddings = torch.reshape(embeddings, tuple(resp.dim)).cuda()
return embeddings
def smis_to_hidden(self, smis):
spec = InputSpec(smis=smis)
resp = self.stub.SmilesToHidden(spec)
hidden_states = torch.FloatTensor(list(resp.hidden_states))
hidden_states = torch.reshape(hidden_states, tuple(resp.dim)).cuda()
masks = torch.BoolTensor(list(resp.masks))
masks = torch.reshape(masks, tuple(resp.dim[:2])).cuda()
return hidden_states, masks
def hidden_to_smis(self, hidden_states, masks):
dim = hidden_states.shape
spec = InputSpec(hidden_states=hidden_states.flatten().tolist(),
dim=dim,
masks=masks.flatten().tolist())
resp = self.stub.HiddenToSmis(spec)
return resp.smis
The wrapper above is meant to condense the workflow shown below. After connecting, initialize a set of SMILES strings which will be encoded.
connection = InferenceWrapper()
smis = ['c1cc2ccccc2cc1',
'COc1cc2nc(N3CCN(C(=O)c4ccco4)CC3)nc(N)c2cc1OC']
Convert SMILES to Embedding#
smis_to_embedding queries the model to fetch the encoder embedding for the input SMILES.
embedding = connection.smis_to_embedding(smis)
embedding.shape
which returns torch.Size([2, 512])
Supplementary Examples with ESM1 and ProtT5#
The inference wrappers for ESM1 and ProtT5 function in a similar manner to MegaMolBART, with the obvious exception of architecture and model function. Full notebooks with examples can be found at bionemo/examples/protein/[esm1nv|prott5]/nbs/Inference.ipynb
The ESM1 and ProtT5 inference wrappers implements seq_to_embedding, which is used to obtain encoder embeddings for the input protein sequence in text format. The batch size, which is the number of sequences submitted at once, may be limited by the compute capacity of the node hosting the model.
From bionemo.examples.protein.[esm1nv|prott5].nbs.infer.py import the inference wrapper. If both wrappers have been imported, it is possible to swap between models.
connection = ESMInferenceWrapper() # // connection = ProtT5nvInferenceWrapper()
seqs = ['MSLKRKNIALIPAAGIGVRFGADKPKQYVEIGSKTVLEHVL', 'MIQSQINRNIRLDLADAILLSKAKKDLSFAEIADGTGLA']
Convert Sequence to Embedding#
The seq_to_embedding method queries the model to fetch the encoder embedding for the input protein sequence.
embeddings = connection.seq_to_embedding(seqs)
embeddings.shape
which returns torch.Size([2, 43, 768])
