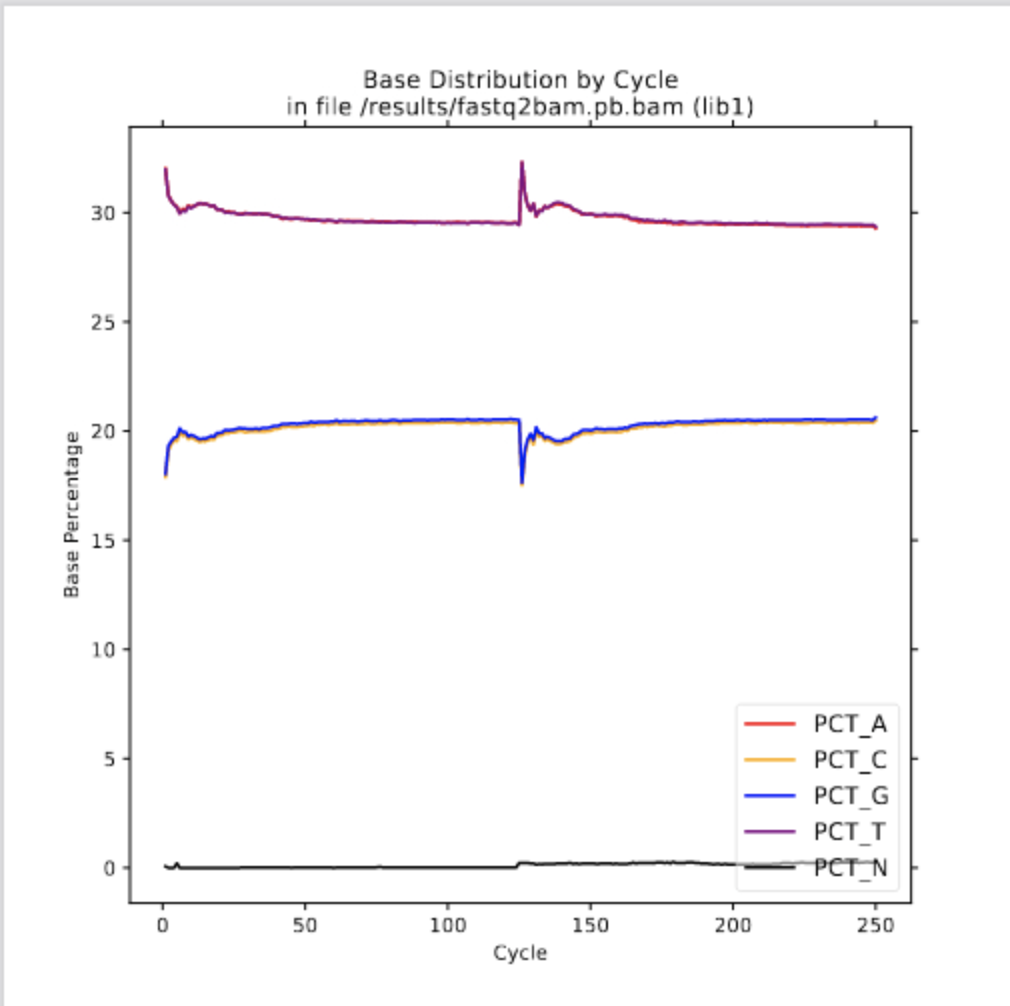
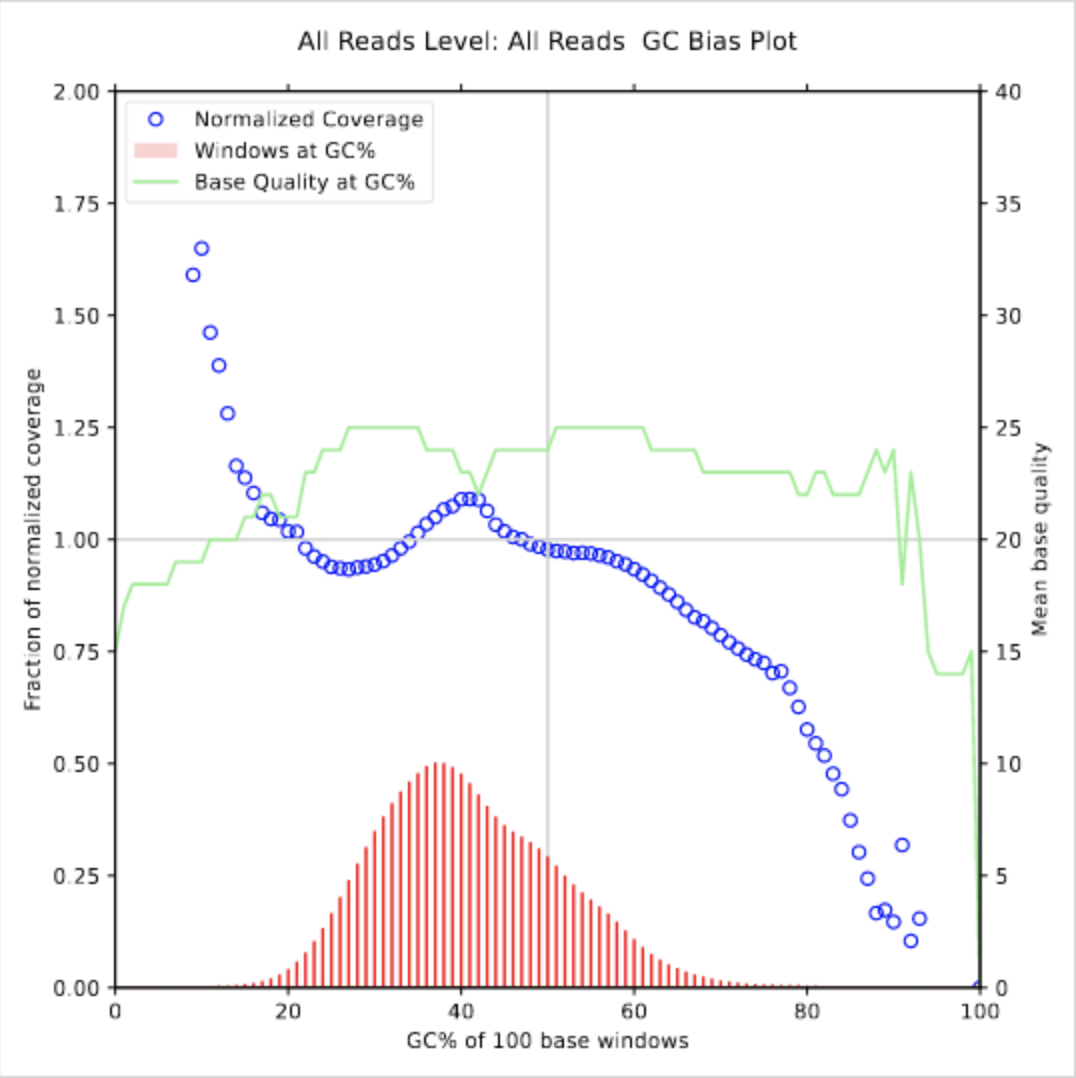
Step #2: Running fq2bam
The following examples will require using the system console of the GPU host. Click on the “System Console” link in the left menu of this page to open a web-based SSH session.

The fq2bam command runs read alignment, sorting, duplicate marking, and base quality score recalibration (BQSR), according to GATK best practices, but at a much faster rate than community tools by leveraging the GPUs.
docker run --gpus all --rm \
-v $(pwd):/results \
-v $(pwd):/data \
-w /data nvcr.io/nvidia/clara/clara-parabricks:4.0.0-1 \
pbrun fq2bam \
--in-fq $FASTQ1 $FASTQ2 \
--ref ${REFERENCE_FILE} \
--out-bam /results/fastq2bam.pb.bam \
--out-qc-metrics-dir qc-metrics
The output should look like:
[Parabricks Options Mesg]: Checking argument compatibility
[Parabricks Options Mesg]: Automatically generating ID prefix
[Parabricks Options Mesg]: Read group created for /data/HG002/MPHG002_S1_L001_R1_001.fastq.gz and
/data/HG002/MPHG002_S1_L001_R2_001.fastq.gz
[Parabricks Options Mesg]: @RG\tID:C6UP4ANXX.1\tLB:lib1\tPL:bar\tSM:sample\tPU:C6UP4ANXX.1
[PB Info 2022-Sep-13 08:16:23] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 08:16:23] || Parabricks accelerated Genomics Pipeline ||
[PB Info 2022-Sep-13 08:16:23] || Version 4.0.0-1 ||
[PB Info 2022-Sep-13 08:16:23] || GPU-BWA mem, Sorting Phase-I ||
[PB Info 2022-Sep-13 08:16:23] ---------------------------------------------------------------------------------
[M::bwa_idx_load_from_disk] read 0 ALT contigs
[PB Warning 2022-Sep-13 08:16:51][ParaBricks/src/pbOpts.cu:316]
WARNING
The system has 186 GB, however recommended RAM with 4 GPU is 196 GB.
The run might not finish or might have less than expected performance.
[PB Info 2022-Sep-13 08:16:51] GPU-BWA mem
[PB Info 2022-Sep-13 08:16:51] ProgressMeter Reads Base Pairs Aligned
[PB Info 2022-Sep-13 08:17:41] 5040000 620000000
[PB Info 2022-Sep-13 08:18:34] 10080000 1260000000
[PB Info 2022-Sep-13 08:19:28] 15120000 1870000000
…
[PB Info 2022-Sep-13 09:35:53] 428400000 53550000000
[PB Info 2022-Sep-13 09:36:38]
GPU-BWA Mem time: 4787.160169 seconds
[PB Info 2022-Sep-13 09:36:38] GPU-BWA Mem is finished.
[main] CMD: /usr/local/parabricks/binaries//bin/bwa mem -Z ./pbOpts.txt /data/Test/GCA_000001405.15_GRCh38_no_alt_analysis_set.fna /data/HG002/MPHG002_S1_L001_R1_001.fastq.gz /data/HG002/MPHG002_S1_L001_R2_001.fastq.gz @RG\tID:C6UP4ANXX.1\tLB:lib1\tPL:bar\tSM:sample\tPU:C6UP4ANXX.1
[main] Real time: 4815.253 sec; CPU: 204261.998 sec
[PB Info 2022-Sep-13 09:36:38] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:36:38] || Program: GPU-BWA mem, Sorting Phase-I ||
[PB Info 2022-Sep-13 09:36:38] || Version: 4.0.0-1 ||
[PB Info 2022-Sep-13 09:36:38] || Start Time: Tue Sep 13 08:16:23 2022 ||
[PB Info 2022-Sep-13 09:36:38] || End Time: Tue Sep 13 09:36:38 2022 ||
[PB Info 2022-Sep-13 09:36:38] || Total Time: 80 minutes 15 seconds ||
[PB Info 2022-Sep-13 09:36:38] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:36:41] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:36:41] || Parabricks accelerated Genomics Pipeline ||
[PB Info 2022-Sep-13 09:36:41] || Version 4.0.0-1 ||
[PB Info 2022-Sep-13 09:36:41] || Sorting Phase-II ||
[PB Info 2022-Sep-13 09:36:41] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:36:41] progressMeter - Percentage
[PB Info 2022-Sep-13 09:36:41] 0.0 0.00 GB
[PB Info 2022-Sep-13 09:36:51] 16.2 1.00 GB
[PB Info 2022-Sep-13 09:37:01] 32.4 1.00 GB
[PB Info 2022-Sep-13 09:37:11] 47.1 1.00 GB
[PB Info 2022-Sep-13 09:37:21] 61.2 1.00 GB
[PB Info 2022-Sep-13 09:37:31] 76.2 1.00 GB
[PB Info 2022-Sep-13 09:37:41] 90.4 1.00 GB
[PB Info 2022-Sep-13 09:37:51] Sorting and Marking: 70.002 seconds
[PB Info 2022-Sep-13 09:37:51] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:37:51] || Program: Sorting Phase-II ||
[PB Info 2022-Sep-13 09:37:51] || Version: 4.0.0-1 ||
[PB Info 2022-Sep-13 09:37:51] || Start Time: Tue Sep 13 09:36:41 2022 ||
[PB Info 2022-Sep-13 09:37:51] || End Time: Tue Sep 13 09:37:51 2022 ||
[PB Info 2022-Sep-13 09:37:51] || Total Time: 1 minute 10 seconds ||
[PB Info 2022-Sep-13 09:37:51] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:37:51] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:37:51] || Parabricks accelerated Genomics Pipeline ||
[PB Info 2022-Sep-13 09:37:51] || Version 4.0.0-1 ||
[PB Info 2022-Sep-13 09:37:51] || Marking Duplicates, BQSR ||
[PB Info 2022-Sep-13 09:37:51] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:37:51] progressMeter - Percentage
[PB Info 2022-Sep-13 09:38:01] 0.0 8.50 GB
[PB Info 2022-Sep-13 09:38:11] 0.0 16.68 GB
…
[PB Info 2022-Sep-13 09:50:11] 100.0 0.00 GB
[PB Info 2022-Sep-13 09:50:18] BQSR and writing final BAM: 746.711 seconds
[PB Info 2022-Sep-13 09:50:18] ---------------------------------------------------------------------------------
[PB Info 2022-Sep-13 09:50:18] || Program: Marking Duplicates, BQSR ||
[PB Info 2022-Sep-13 09:50:18] || Version: 4.0.0-1 ||
[PB Info 2022-Sep-13 09:50:18] || Start Time: Tue Sep 13 09:37:51 2022 ||
[PB Info 2022-Sep-13 09:50:18] || End Time: Tue Sep 13 09:50:18 2022 ||
[PB Info 2022-Sep-13 09:50:18] || Total Time: 12 minutes 27 seconds ||
[PB Info 2022-Sep-13 09:50:18] ---------------------------------------------------------------------------------
/tmp/7QB5DKRM_run.sh
Generating qualityscore pdf...
Generating insertsize pdf...
Generating meanqualitybycycle pdf...
Generating qualityscore pdf...
Generating gcbias pdf...
Please visit https://docs.nvidia.com/clara/#parabricks for detailed documentation
Now let’s look at the outputs generated :
ls -l fastq2bam*
-rw-r--r-- 1 root root 4819386784 Sep 6 12:43 fastq2bam.pb.bam
-rw-r--r-- 1 root root 6882792 Sep 6 12:43 fastq2bam.pb.bam.bai
ls qc-metrics/
alignment.txt insert_size.pdf qualityscore.png
base_distribution_by_cycle.pdf insert_size.png qualityscore.txt
base_distribution_by_cycle.png insert_size.txt sequencingArtifact.bait_bias_detail_metrics.txt
base_distribution_by_cycle.txt mean_quality_by_cycle.pdf sequencingArtifact.bait_bias_summary_metrics.txt
gcbias.pdf mean_quality_by_cycle.png sequencingArtifact.error_summary_metrics.txt
gcbias_0.png mean_quality_by_cycle.txt sequencingArtifact.pre_adapter_detail_metrics.txt
gcbias_detail.txt quality_yield.txt sequencingArtifact.pre_adapter_summary_metrics.txt
gcbias_summary.txt qualityscore.pdf