Pipeline Tools
In Clara Parabricks each pipeline is a collection of several individual tools that are commonly used together, all wrapped up as a single tool. For example, the deepvariant_germline takes FASTA and FASTQ files as input and produces a VCF and BAM file as output. Internally, it runs BWA mem alignment, performs coordinate sorting, marks duplicates, and then runs DeepVariant.
Clara Parabricks contains the following pipelines. Click on the name of a pipeline for tool-specific options.
Tool |
Details |
Run the germline pipeline from FASTQ to VCF using a deep neural network analysis |
|
Run the germline pipeline from FASTQ to VCF |
|
Run the somatic pipeline from FASTQ to VCF |
While the user cannot alter or create their own Clara Parabricks pipeline, Parabricks containers are compatible with WDL and NextFlow for building customized workflows, intertwining GPU- and CPU-powered tasks with different compute requirements, and deploying at scale.
These enable workflows to be deployed on cloud batch services as well as local clusters (e.g. SLURM) in a well managed process, pulling from a combination of Clara Parabricks and third-party containers and running these on pre-defined nodes.
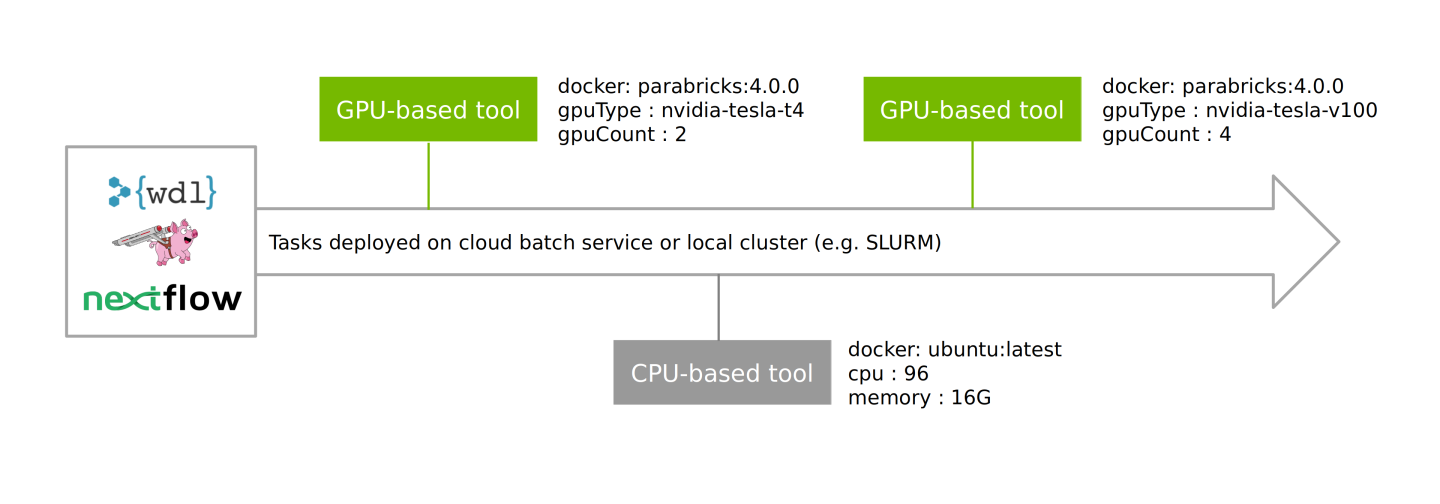
For further information on running these workflows, and to see the open-source reference workflows, which can be easily forked/edited, visit the Clara Parabricks Workflows repository. This repository includes recommended instance configurations for deploying the GPU-based tools on cloud and can be easily forked/edited for your own purposes.