- What's New?
- Getting Started with Clara Parabricks
- Software Overview
- Tutorials
- How-Tos
- Tool Reference
- Help
- References
How the Documentation is Organized
Introduction: This is the main page, which contains a brief introduction to Clara Parabricks: What it is, what it can do, and how it works.
What's New? covers what's changed since the previous release: new tools, improvements to existing tools, and bug fixes.
Getting Started with Clara Parabricks page focuses on all the steps of setting up the software, including requirements, examples and optimizing it for performance
Tutorials walk you through a single use of Clara Parabricks using an example dataset. The steps will familiarize the users with the software and walk you through a reproducible example. It will start from a reference and FASTQ files to a BAM file, then do variant calling on the BAM file, and produce a VCF file.
How-Tos explore larger, more involved tasks, examining a wider variety of options, tools, and workflows. Owing to the larger data sets in use, a more capable hardware platform may be required (more GPUs, more memory, etc).
Tool Reference contains reference documentation for each tool, organized both by category and alphabetically by tool name. It also tells users how to compare the output of Clara Parabricks with the output from the baseline tools. A list of publications referencing Clara Parabricks, a list of frequently asked question, and pointers on getting more help and information are also part of this section
What is Clara Parabricks?
Parabricks is a software suite for performing secondary analysis of next generation sequencing (NGS) DNA and RNA data. It delivers results at blazing fast speeds and low cost. Clara Parabricks can analyze 30x WGS (whole human genome) data in about 25 minutes, instead of 30 hours for other methods. Its output matches commonly used software, making it fairly simple to verify the accuracy of the output.
Why use Clara Parabricks?
Under the hood, Parabricks achieves this performance through tight integration with GPUs, which excel at performing data-parallel computation much more effectively than traditional CPU-based solutions. Parabricks was built from the ground up by GPU computing and Deep Learning experts who wanted to develop the fastest and most efficient possible implementation of common genomics algorithms used in secondary analysis.
Learn more at the Clara Parabricks developer page.
Software Overview
Parabricks is a software suite for genomic analysis. It delivers major improvements in throughput time for common analytical tasks in genomics, including germline and somatic analysis. The core of the Parabricks software is its data pipeline, which takes raw data and transforms it according to the user's requirements.
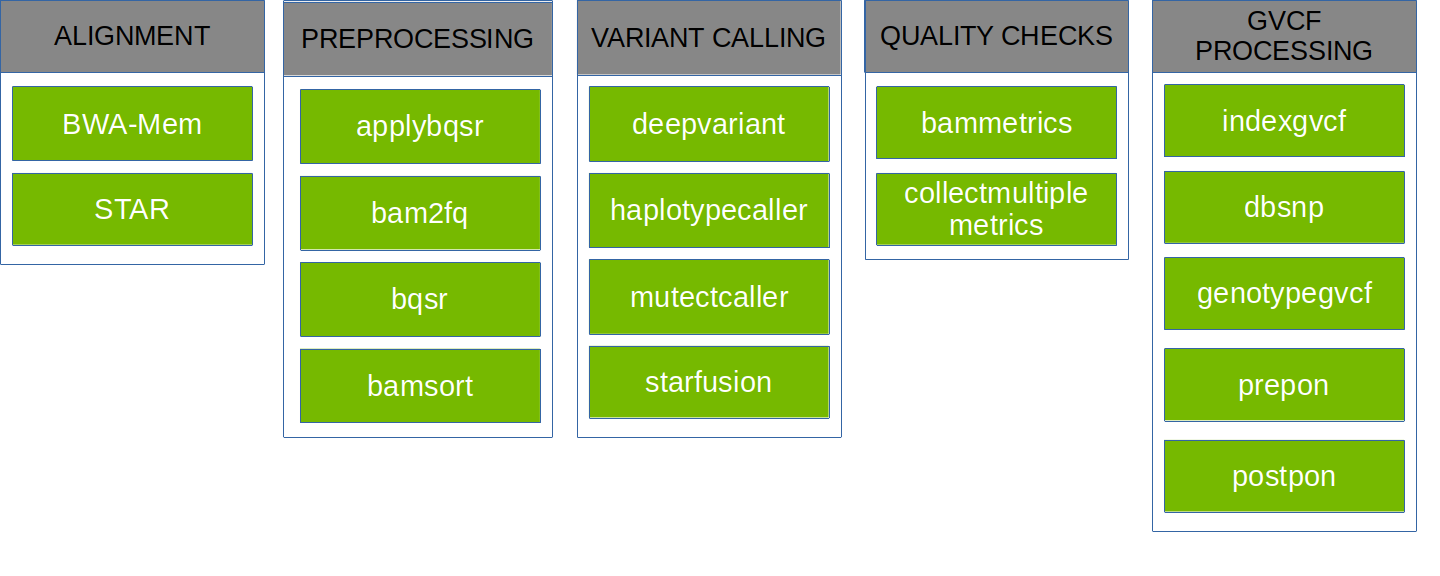
The Parabricks software supports the tools shown below:
Parabricks Pipelines
Parabricks pipelines can simplify the steps required for processing. In Clara Parabricks, each pipeline is a collection of several individual tools that are commonly used together, all wrapped up as a single tool. For example, the deepvariant pipeline takes FASTA and FASTQ files as input and produces a VCF and BAM file as output. Internally, it runs BWA mem alignment, performs coordinate sorting, marks duplicates, and then runs DeepVariant.
Parabricks supports the pipelines shown below:
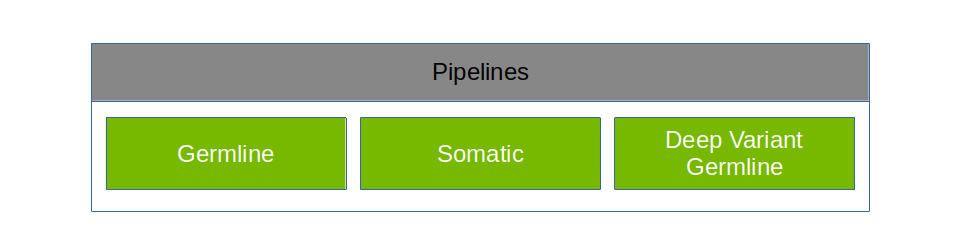
Parabricks can be configured to run specific accelerated tools or run full pipelines that are commonly used. The Software Tools section covers individual tools, and the Pipelines section covers commonly used pipelines.
NVIDIA Parabricks pipelines have been tested on Dell, HPE, IBM, and NVIDIA servers at Amazon Web Services, Google Cloud, Oracle Cloud Infrastructure, and Microsoft Azure.
How to Get Help
For technical support, updated user guides, and other Parabricks documentation, go to https://docs.nvidia.com/clara/#parabricks.
Answers to most FAQs can be found on the developer forum at https://forums.developer.nvidia.com/c/healthcare/Parabricks/290.