Overview
NVIDIA co-founded Project MONAI, the Medical Open Network for AI, with the world’s leading academic medical centers to establish an inclusive community of AI researchers to develop and exchange best practices for AI in healthcare imaging across academia and enterprise researchers.
MONAI is the domain-specific, open-source Medical AI framework that drives research breakthroughs and accelerates AI into clinical impact. MONAI unlocks the power of medical data to build deep learning models for medical AI workflows. MONAI provides the essential domain-specific tools from data labeling to model training, making it easy to develop, reproduce and standardize medical AI lifecycles.
MONAI Enterprise is NVIDIA’s offering for enterprise-grade use of MONAI with an NVIDIA AI Enterprise 3.0 license. MONAI Enterprise on NVIDIA AI Enterprise 3.0 offers the MONAI Toolkit container, which provides enterprise developers and researchers with a secure, scalable workflow to develop medical imaging AI.
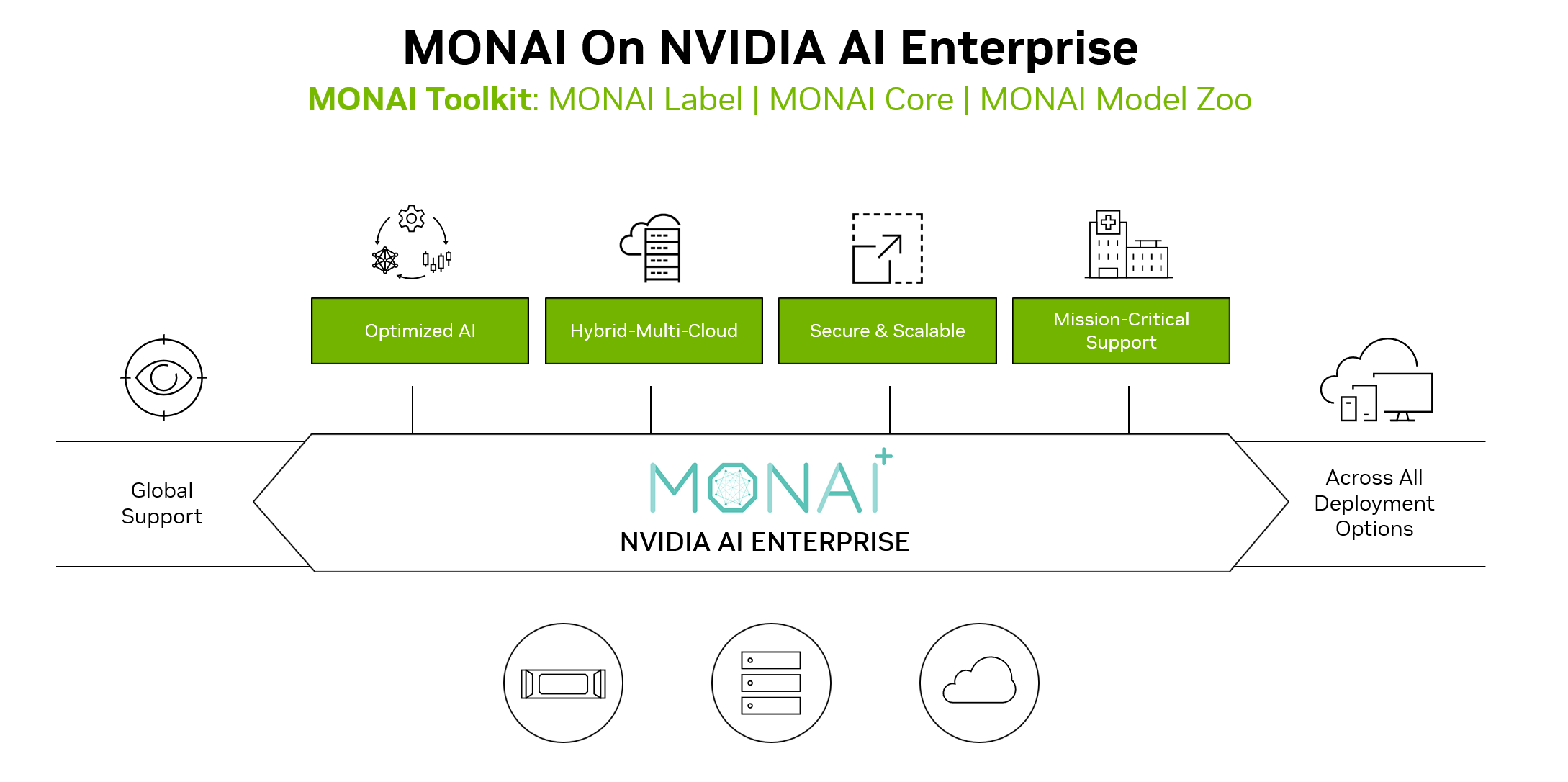
MONAI Toolkit is a development sandbox offered as part of MONAI Enterprise, an NVIDIA AI Enterprise-supported distribution of MONAI. It includes a base container and a curated library of 9 pre-trained models (CT, MR, Pathology, Endoscopy), available on NGC, that allows data scientists and clinical researchers to jumpstart AI development. The base container includes the following:
MONAI Label: An intelligent labeling and learning tool with active learning that reduces data labeling costs by 75%
MONAI Core: A training framework to build robust AI models with self-supervised learning, federated learning, and Auto3D segmentation.
With federated learning, APIs algorithms built with NVIDIA FLARE, MONAI can run on any federated learning platform.
Auto3DSeg is domain-specialized AutoML for 3D segmentation, accelerating the development of medical imaging models and maximizing researcher productivity and throughput. Developers can get started with 1-5 lines of code, reducing training time from weeks/months to 2 days.
MONAI Model Zoo: A curated library of 9 pre-trained models (CT, MR, Pathology, Endoscopy) that allows data scientists and clinical researchers to jumpstart AI development
Curated Jupyter notebooks and tutorial resources to ease the onboarding process.
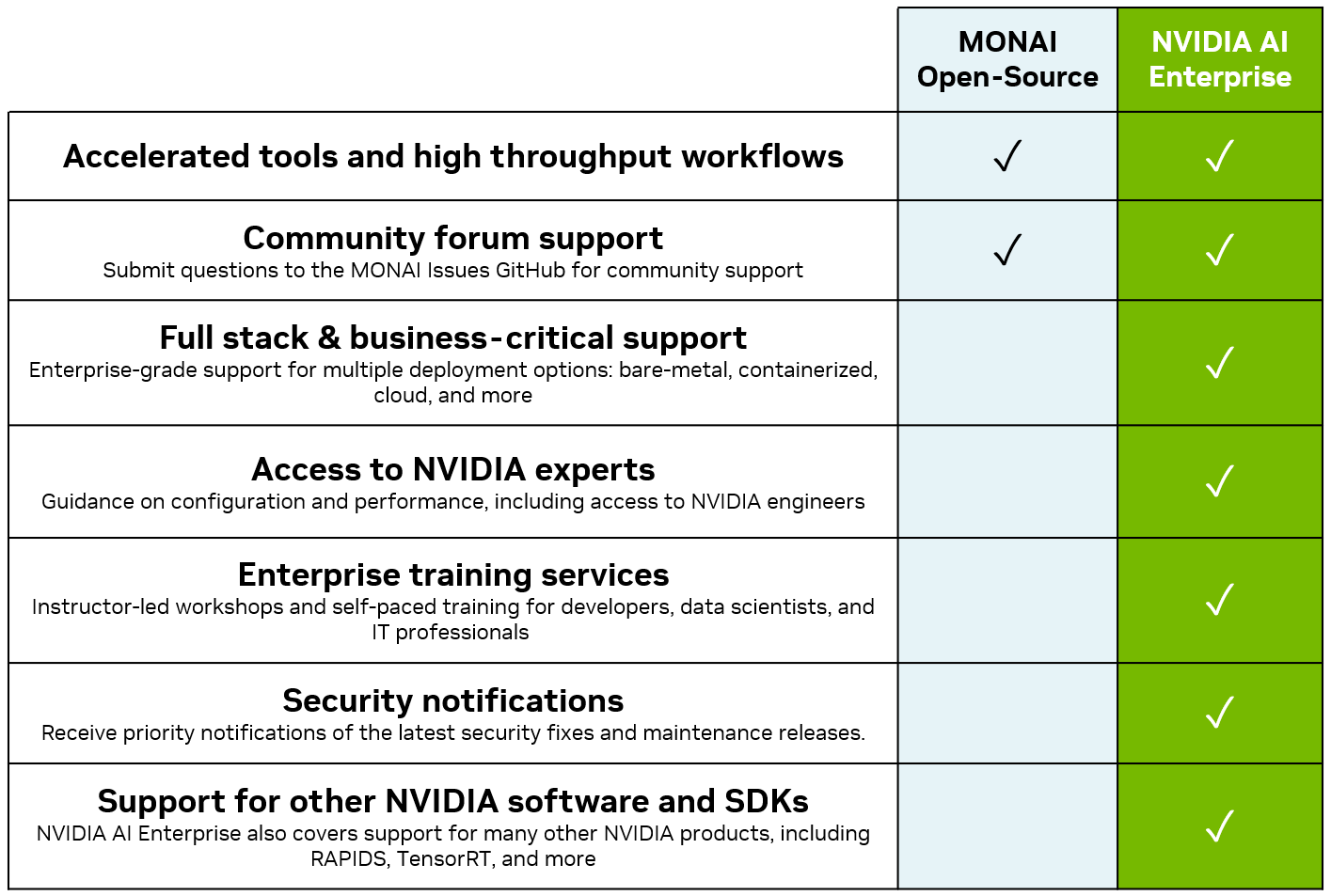
Table of Contents
System Requirements
Installation Prerequisites
Using the MONAI Toolkit Container requires the host system to have the following installed:
The MONAI Toolkit Container uses the 22.09 base container image.
For a full list of supported software and specific versions that come packaged with the frameworks based on the container image, see the Framework Containers Support Matrix and the NVIDIA Container Toolkit Documentation.
No other installation, compilation, or dependency management is required. It is not necessary to install the NVIDIA CUDA Toolkit.
Hardware Requirements
MONAI Toolkit has been validated on V100 and A100 GPUs, but should generally be supported on GPUs such as H100, A100, A40, A30, A10, V100, and more.
The following system configuration is recommended to achieve reasonable training and inference performance with MONAI Toolkit and supported models provided.
At least 12GB of GPU RAM and up to 32GB depending on the network or model you use
At least 32 GB of RAM - and the more RAM the better
At least 50 GB of SSD Disk Space
For a complete list of supported hardware, please see the NVIDIA AI Enterprise Product Support Matrix
Who is this toolkit for?
Researcher
A Deep Learning Researcher’s role is to build novel domain-specific models and techniques to drive AI-powered acceleration for workflows or products. MONAI allows researchers to utilize standard building blocks to focus on creating their state-of-the-art AI models.
Data Scientist
A Data Scientist is a user/consumer of a research pipeline. The data scientist’s role is to adapt existing state-of-the-art domain techniques for specific data and use cases. They’ll refine and fine-tune the data science pipeline to fit the clinical workflow/application needed. MONAI aims to be easily integrated into existing pipelines, whether you already have a PyTorch workflow or you’re just getting started on your task.
Application Developer
A platform developer aims to build a platform and service leveraging MONAI components that may include:
A labeling or active learning platform using MONAI Label
A complete ML platform for data scientists, integrated with data stores for data flow management with MONAI Label and Core.
IT Admin
An IT admin’s role is to provision and configure systems to stand up the MONAI Toolkit sandbox to serve an enterprise’s data science and research teams. The toolkit provides the perfect base for those teams to get started quickly with guided tutorials and validated workflows.
Toolkit Libraries
Software Versions
| Software | Version |
| ----------- | ----------- |
| MONAI Core | 1.0.1 |
| MONAI Label | 0.5.2 |
| NVFlare | 2.2.1 |
MONAI Label
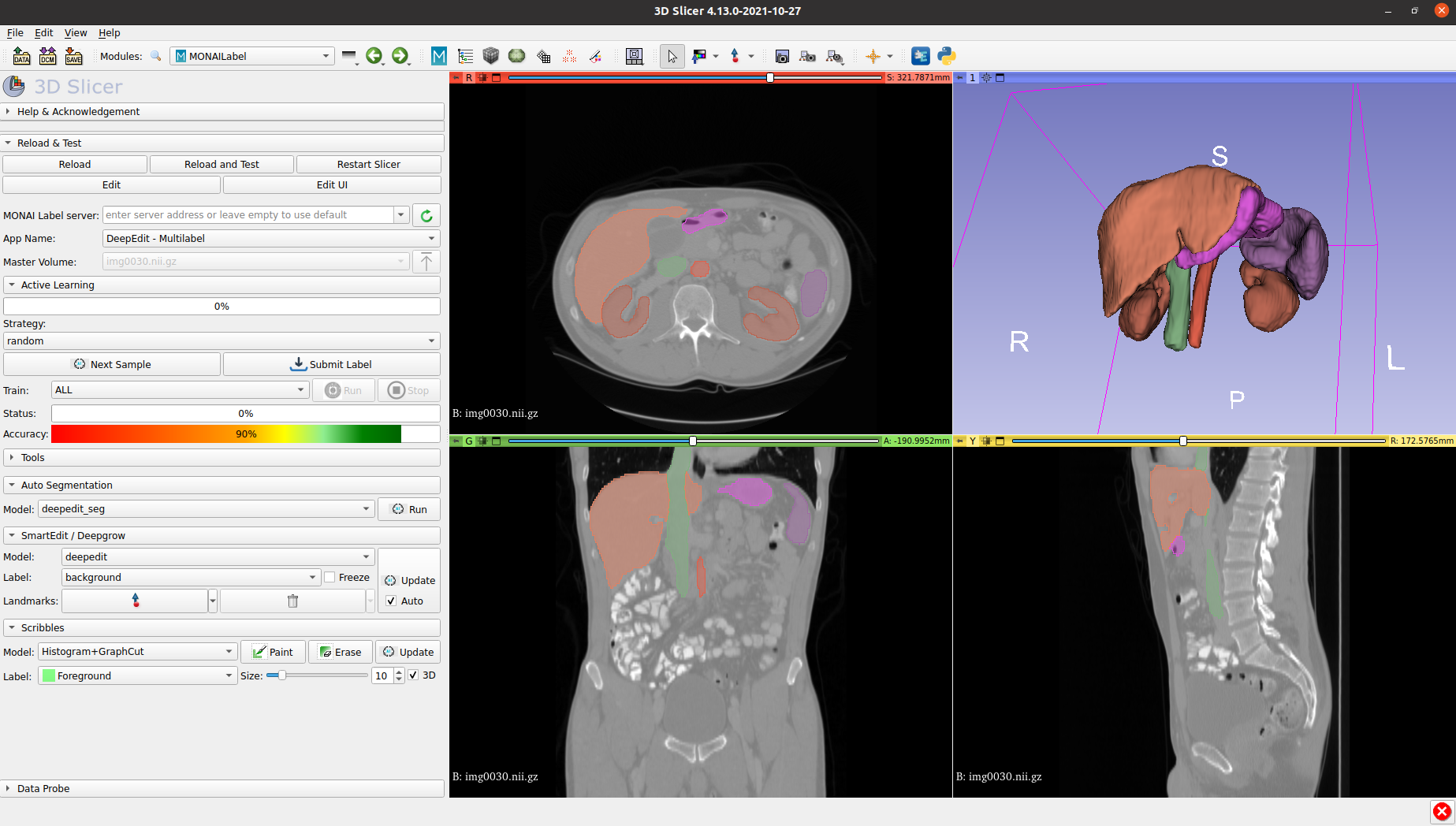
MONAI Label minimizes the need for developers and researchers to manually annotate data by providing an intelligent imaging labeling and learning SDK that can be integrated into either customized or template-based data labeling apps. MONAI Label saves developers time, helping them quickly progress to training, tuning, and validating their medical AI models within the MONAI standardized paradigm.
The industry’s most popular open source viewers already have MONAI integrated: 3D Slicer, OHIF, QuPath, Digital Slide Archive, and CVAT, and it is also integrated into cloud service providers. Developers can also incorporate MONAI Label into their custom viewer using server and client APIs, which are well abstracted and documented for seamless integration.
MONAI Label has enhanced active learning capabilities, which is a process that aims to use the least amount of data to achieve the highest possible model performance. Choosing data that will have the most significant influence on your overall model accuracy allows human annotators to focus on the annotations that will have the highest impact on the model performance. Developers can see up to a 75% reduction in training costs with active learning in MONAI Label with increased labeling and training efficiency while achieving better model performance.
MONAI Core
MONAI Core gives developers and researchers a PyTorch-driven library for deep learning tasks that includes domain-optimized capabilities they need for developing medical imaging training workflows. Performance features such as MONAI Core’s AutoML, Smart Caching, GPU-accelerated I/O, and transforms take training from days to hours, and hours to minutes, helping users accelerate AI into clinical production.
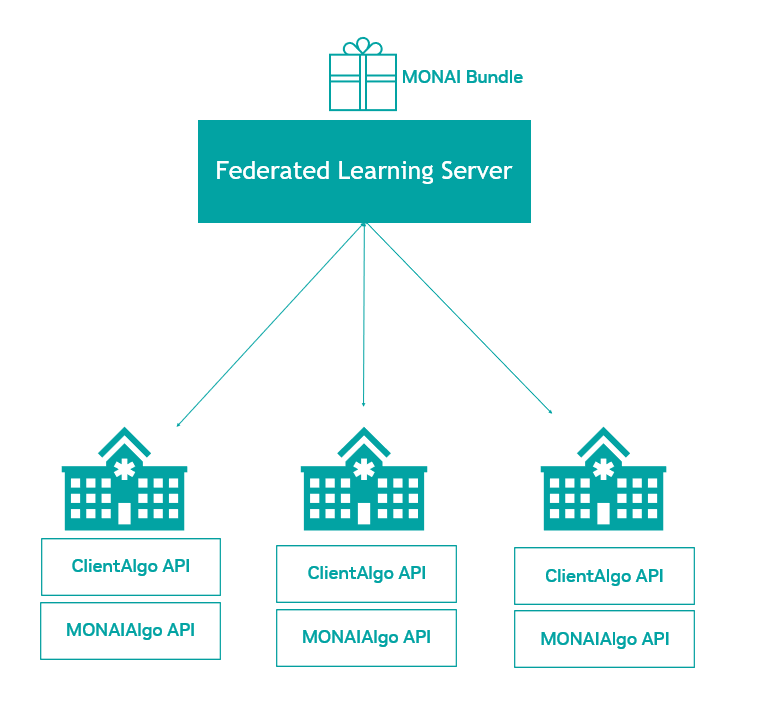
With Auto3D segmentation, developers can train models with 1-5 lines of code that can quickly segment regions of interest in data from 3D imaging modalities like CT and MRI, reducing training time from weeks/months to 2 days. And MONAI Core’s federated learning client algorithm APIs are exposed as an abstract base class for defining an algorithm to run on any federated learning platform. NVIDIA FLARE has already built the integration piece with these new APIs. And using MONAI Bundle configurations with the new federated learning APIs means developers can take any bundle and seamlessly extend them to a federated paradigm.
NVIDIA FLARE
NVIDIA FLARE (NVIDIA Federated Learning Application Runtime Environment) is a domain-agnostic, open-source, extensible SDK that allows researchers and data scientists to adapt existing ML/DL workflows (PyTorch, RAPIDS, Nemo, TensorFlow) to a federated paradigm. It enables platform developers to build a secure, privacy-preserving offering for a distributed multi-party collaboration. Built on a componentized architecture, NVIDIA FLARE gives you the flexibility to take federated learning workloads from research and simulation to real-world production deployment.
MONAI includes the federated learning client algorithm APIs exposed as an abstract base class for defining an algorithm to run on any federated learning platform. NVIDIA FLARE has already built the integration piece with these new APIs.
Quickstart Guide
Usage
Running the MONAI Toolkit JupyterLab Instance
To start with a local host system, the MONAI Toolkit JupyterLab instance can be started with a ready-to-open website link with:
docker run --gpus all -it --rm --ipc=host --net=host nvcr.io/nvidia/clara/monai-toolkit /opt/docker/runtoolkit.sh
After the JupyterLab App is started, follow the onscreen instruction and open the URL in a web browser.
Note: By default, the container use the host system’s all GPU resources, networking and inter-process communication (IPC) namespace. Multiple notebooks require a large shared memory size for the container to run comprehensive workflows. For more information, please refer to changing the shared memory segment size
Running the MONAI Toolkit in Interactive Bash
To run the MONAI Toolkit container with the bash shell, issue the command below to start the prebuilt container:
docker run --gpus all -it --rm --ipc=host --net=host \
nvcr.io/nvidia/clara/monai-toolkit \
/bin/bash
Changing Shared Memory Segment Size
DIGITS uses shared memory to share data between processes. For example, if you use Torch multiprocessing for multi-threaded data loaders, the default shared memory segment size that the container runs with may not be enough. Therefore, you should increase the shared memory size by issuing either:
--ipc=host
or
--shm-size=
Access the JupyterLab Remotely
Users can access the JupyterLab instance by simply typing the host machine’s URL or IP address in the web browser with the port number (default: 8888).
A token may be required to log in for the first time.
Users can find the token in the system which hosts the MONAI Toolkit container by looking for the code after /?token= on the screen.
Running the JupyterLab Instance on a Remote Host Machine
JupyterLab is started on port 8888 by default. If the user wants to assign another port, the JupyterLab instance can be started by setting the JUPYTER_PORT environment variable:
-e JUPYTER_PORT=8900
For example:
docker run --gpus all -it --rm --ipc=host --net=host \
-e JUPYTER_PORT=8900 \
nvcr.io/nvidia/clara/monai-toolkit \
/opt/docker/runtoolkit.sh
Note: More details about running docker commands are explained in the Running A Container chapter in the NVIDIA Containers For Deep Learning Frameworks User’s Guide and specify the registry, repository, and tags.
Mount Custom Data Directory
To mount a custom data directory, the users can use -v to mount the drive(s) and override the default data directory environment variable MONAI_DATA_DIRECTORY used by many notebooks.
For example:
docker run --gpus all -it --rm --ipc=host --net=host \
-v ~/workspace:/workspace \
-e MONAI_DATA_DIRECTORY=/workspace/data \
nvcr.io/nvidia/clara/monai-toolkit \
/opt/docker/runtoolkit.sh
Configure JupyterLab
/opt/docker/runtoolkit.sh provides an entry point for the user to configure Jupyter Lab with the default settings.
However, it cannot cover all use scenarios and APIs of Jupyter.
The user can run the Jupyter Lab command in the MONAI Toolkit container and configure the instance simply by:
docker run --gpus all -it --rm --ipc=host --net=host nvcr.io/nvidia/clara/monai-toolkit jupyter lab
Navigating the MONAI Toolkit
We recommend going sequentially through each chapter and focusing on the specific types of tasks that you’re interested in (e.g., Radiology, Pathology, or Computer-Assisted Intervention).
For additional information on setting up your environment and using the MONAI Toolkit, check out Chapter 1: Getting Started.
If you are at the start of your journey and looking to understand how to speed up your image annotation process, check out Chapter 2: Using MONAI Label.
If you already have an annotated dataset and want to get started training with MONAI or integrate MONAI into your existing PyTorch Training Loop, take a Chapter 3: Using MONAI Core.
If you are interested in MONAI and Federated Learning working together, look at Chapter 4: MONAI Federated Learning.
If you are interested in some advanced topics and looking to compare benchmarks, interested in performance profiling, or looking for some researcher best practices with MONAI, check out Chapter 5: Performance and Benchmarking.
Additional Resources
Below you can find additional resources on MONAI and NVIDIA.