deepvariant_germline
Given one or more pairs of FASTQ files, you can run the germline variant pipeline workflow to generate output, including variants, BAM, and recal.
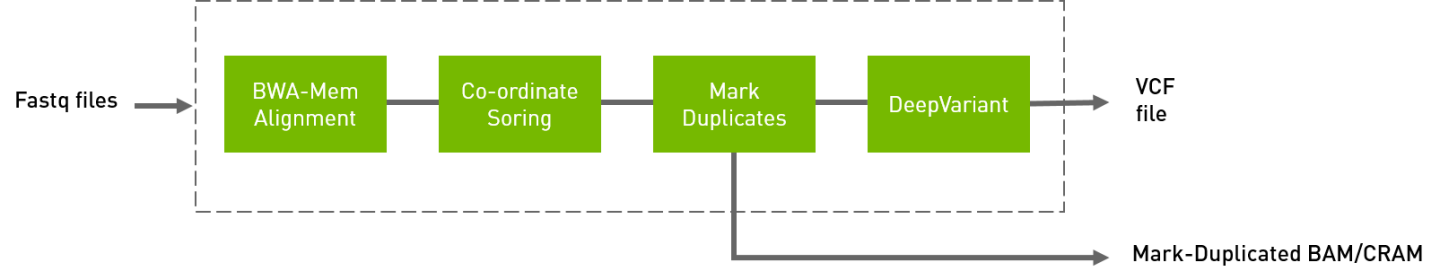
The deepvariant germline pipeline includes alignment, sorting, and marking, as well as the deepvariant variant caller.
Currently, Deepvariant is supported for T4, V100, and A100 GPUs only.
The inputs are BWA-indexed reference files and pair-ended FASTQ files. The outputs of this pipeline are the following:
Aligned, co-ordinate sorted, duplicated marked BAM
Variants in
vcf/g.vcf/g.vcf.gzformat
The following command runs the DeepVariant pipeline.
$ pbrun deepvariant_germline \
--ref Ref/Homo_sapiens_assembly38.fasta \
--in-fq input1.fq input2.fq \
--out-variants variants.vcf
Run the germline pipeline from FASTQ to VCF using a deep neural network analysis.
Input/Output file options
- --ref REF
- --in-fq [IN_FQ [IN_FQ ...]]
- --in-se-fq [IN_SE_FQ [IN_SE_FQ ...]]
- --knownSites KNOWNSITES
- --interval-file INTERVAL_FILE
- --pb-model-file PB_MODEL_FILE
- --out-recal-file OUT_RECAL_FILE
- --out-bam OUT_BAM
- --out-variants OUT_VARIANTS
- --out-duplicate-metrics OUT_DUPLICATE_METRICS
Path to the reference file. (default: None)
Option is required.
Path to the pair ended FASTQ files followed by optional read group with quotes (Example: "@RGtID:footLB:lib1tPL:bartSM:sampletPU:foo"). Files must be in fastq/fastq.gz format. All sets of inputs should have a read group; otherwise, none should have a read group, and it will be automatically added by the pipeline. This option can be repeated multiple times. Example 1: --in-fq sampleX_1_1.fastq.gz sampleX_1_2.fastq.gz --in-fq sampleX_2_1.fastq.gz sampleX_2_2.fastq.gz . Example 2: --in-fq sampleX_1_1.fastq.gz sampleX_1_2.fastq.gz "@RG ID:footLB:lib1tPL:bartSM:sampletPU:unit1" --in-fq sampleX_2_1.fastq.gz sampleX_2_2.fastq.gz "@RG ID:foo2tLB:lib1tPL:bartSM:sampletPU:unit2" . For same sample, Read Groups should have same sample name (SM) and different ID and PU. (default: None)
Path to the single ended FASTQ file followed by optional read group with quotes (Example: "@RGtID:footLB:lib1tPL:bartSM:sampletPU:foo"). The file must be in fastq/fastq.gz format. All sets of inputs should have a read group; otherwise, none should have a read group, and it will be automatically added by the pipeline. This option can be repeated multiple times. Example 1: --in-se-fq sampleX_1.fastq.gz --in-se-fq sampleX_2.fastq.gz . Example 2: --in-se-fq sampleX_1.fastq.gz "@RGtID:footLB:lib1tPL:bartSM:sampletPU:unit1" --in-se-fq sampleX_2.fastq.gz "@RGtID:foo2tLB:lib1tPL:bartSM:sampletPU:unit2" . For same sample, Read Groups should have same sample name (SM) and different ID and PU. (default: None)
Path to a known indels file. Must be in vcf/vcf.gz format. This option can be used multiple times. (default: None)
Path to an interval file with possible formats: Picard-style (.interval_list or .picard), GATK-style (.list or .intervals), or BED file (.bed). This option can be used multiple times. (default: None)
Path of non-default parabricks model file for deepvariant. (default: None)
Path of the report file after Base Quality Score Recalibration. (default: None)
Path of BAM file after Marking Duplicates. (default: None)
Option is required.
Path of the vcf/gvcf/gvcf.gz file after variant calling. (default: None)
Option is required.
Path of a duplicate metrics file after Marking Duplicates. (default: None)
Options specific to this tool
- -L INTERVAL, --interval INTERVAL
- --bwa-options BWA_OPTIONS
- --no-warnings
- --no-markdups
- --fix-mate
- --markdups-assume-sortorder-queryname
- --markdups-picard-version-2182
- --optical-duplicate-pixel-distance OPTICAL_DUPLICATE_PIXEL_DISTANCE
- --read-group-sm READ_GROUP_SM
- --read-group-lb READ_GROUP_LB
- --read-group-pl READ_GROUP_PL
- --read-group-id-prefix READ_GROUP_ID_PREFIX
- --disable-use-window-selector-model
- --gvcf
- --norealign-reads
- --sort-by-haplotypes
- --keep-duplicates
- --vsc-min-count-snps VSC_MIN_COUNT_SNPS
- --vsc-min-count-indels VSC_MIN_COUNT_INDELS
- --vsc-min-fraction-snps VSC_MIN_FRACTION_SNPS
- --vsc-min-fraction-indels VSC_MIN_FRACTION_INDELS
- --min-mapping-quality MIN_MAPPING_QUALITY
- --min-base-quality MIN_BASE_QUALITY
- --mode MODE
- --alt-aligned-pileup ALT_ALIGNED_PILEUP
- --variant-caller VARIANT_CALLER
- --add-hp-channel
- --parse-sam-aux-fields
- --use-hp-information
- --use-wes-model
Interval within which to call bqsr from the input reads. All intervals will have a padding of 100 to get read records and overlapping intervals will be combined. Interval files should be passed using the --interval-file option. This option can be used multiple times. e.g. "-L chr1 -L chr2:10000 -L chr3:20000+ -L chr4:10000-20000" (default: None)
Pass supported bwa mem options as one string. Current original bwa mem supported options, -M, -Y, -T. e.g. --bwa-options="-M -Y" (default: None)
Suppress warning messages about system thread and memory usage (default: None)
Do not perform Mark Duplicates step. Return BAM after sorting. (default: None)
Add mate cigar (MC) and mate quality (MQ) tags to the output file. (default: None)
Assume the reads are sorted by queryname for Marking Duplicates. This will mark secondary, supplementary and unmapped reads as duplicates as well. This flag will not impact variant calling while increasing processing times (default: None)
Assume marking duplicates to be similar to Picard version 2.18.2 (default: None)
The maximum offset between two duplicate clusters in order to consider them optical duplicates. Ignored if --out-duplicate-metrics is not passed (default: None)
SM tag for read groups in this run (default: None)
LB tag for read groups in this run (default: None)
PL tag for read groups in this run (default: None)
prefix for ID and PU tag for read groups in this run. This prefix will be used for all pair of fastq files in this run. The ID and PU tag will consist of this prefix and an identifier which will be unique for a pair of fastq files (default: None)
Change the window selector model from Allele Count Linear to Variant Reads. This option will increase the accuracy and runtime. (default: None)
Generate variant calls in gvcf format. (default: None)
Do not locally realign reads before calling variants. Reads longer than 500 base pairs are never realigned. (default: None)
Reads are sorted by haplotypes (using HP tag) (default: None)
Keep reads that are duplicate. (default: None)
SNP alleles occurring at least this many times in our AlleleCount will be advanced as candidates. (default: 2)
Indel alleles occurring at least this many times in our AlleleCount will be advanced as candidates. (default: 2)
SNP alleles occurring at least this fraction of all counts in our AlleleCount will be advanced as candidates. (default: 0.12)
Indel alleles occurring at least this fraction of all counts in our AlleleCount will be advanced as candidates. (default: None)
By default, reads with any mapping quality are kept. Setting this field to a positive integer i will only keep reads that have a MAPQ >= i. Note this only applies to aligned reads. (default: 5)
Minimum base quality. This field indicates that we are enforcing a minimum base quality score for alternate alleles. Alternate alleles will only be considered if all bases in the allele have a quality greater than min_base_quality. (default: 10)
Value can be one of [shortread, pacbio, ont]. By default, it is shortread. If mode is set to pacbio, the following defaults are used: --norealign-reads, --alt-aligned-pileup diff_channels, --vsc-min-fraction-indels 0.12. If mode is set to ont, the following defaults are used: -norealign-reads, --variant-caller VCF_CANDIDATE_IMPORTER. (default: shortread)
Value can be one of [none, diff_channels]. Include alignments of reads against each candidate alternate allele in the pileup image. Default is none which turns this feature off. (default: None)
Value can be one of [VERY_SENSITIVE_CALLER, VCF_CANDIDATE_IMPORTER]. The caller to use to make examples. If you use VCF_CANDIDATE_IMPORTER, it implies force calling. Default is VERY_SENSITIVE_CALLER (default: None)
Add another channel to represent HP tags per read. (default: None)
Auxiliary fields of the BAM/CRAM records are parsed. Needed by --sort-by-haplotypes by --add-hp-channel. (default: None)
If passed, both --sort-by-haplotypes and --parse-sam-aux-fields will be set to true, only used in PACBIO mode. (default: None)
If passed, WES model file will be used. Only used in shortread mode. (default: None)
Common options:
- --logfile LOGFILE
- --tmp-dir TMP_DIR
- --with-petagene-dir WITH_PETAGENE_DIR
- --keep-tmp
- --license-file LICENSE_FILE
- --no-seccomp-override
- --version
Path to the log file. If not specified, messages will only be written to the standard error output. (default: None)
Full path to the directory where temporary files will be stored.
Full path to the PetaGene installation directory. By default, this should have been installed at /opt/petagene. Use of this option also requires that the PetaLink library has been preloaded by setting the LD_PRELOAD environment variable. Optionally set the PETASUITE_REFPATH and PGCLOUD_CREDPATH environment variables that are used for data and credentials (default: None)
Do not delete the directory storing temporary files after completion.
Path to license file license.bin if not in the installation directory.
Do not override seccomp options for docker (default: None).
View compatible software versions.
GPU options:
- --num-gpus NUM_GPUS
- --gpu-devices GPU_DEVICES
Number of GPUs to use for a run. GPUs 0..(NUM_GPUS-1) will be used.
GPU devices to use for a run. By default, all GPU devices will be used.
To use specific GPU devices, enter a comma-separated list of GPU device
numbers. Possible device numbers can be found by examining the output of
the nvidia-smi command. For example, using --gpu-devices 0,1
would only use the first two GPUs.
The --in-fq option takes the names of two FASTQ files, optionally followed by a quoted read group. The FASTQ filenames must not start with a hyphen.