Running NVIDIA Parabricks on nf-core
This guide shows how to run Parabricks on nf-core.
Parabricks is an accelerated compute framework that supports applications across the genomics industry, primarily supporting analytical workflows for DNA, RNA, and somatic mutation detection applications. With industry leading compute times, Parabricks rapidly converts a FASTQ file to a VCF using multiple, industry validated variant callers and also includes the ability to QC and annotate those variants. As Parabricks is based upon publicly available tools, results are easy to verify and combine with other publicly available data sets.
More information is available on the Parabricks Product Page.
Detailed installation, usage, and tuning information is available in the Parabricks user guide.
Several Parabricks pipelines can be found on nf-core as modules. These modules can be dropped into existing workflows or used on their own. To read more about nf-core modules, see their documentation.
To find the available Parabricks modules, visit the nf-core modules page and search for “Parabricks”. The following modules are available:
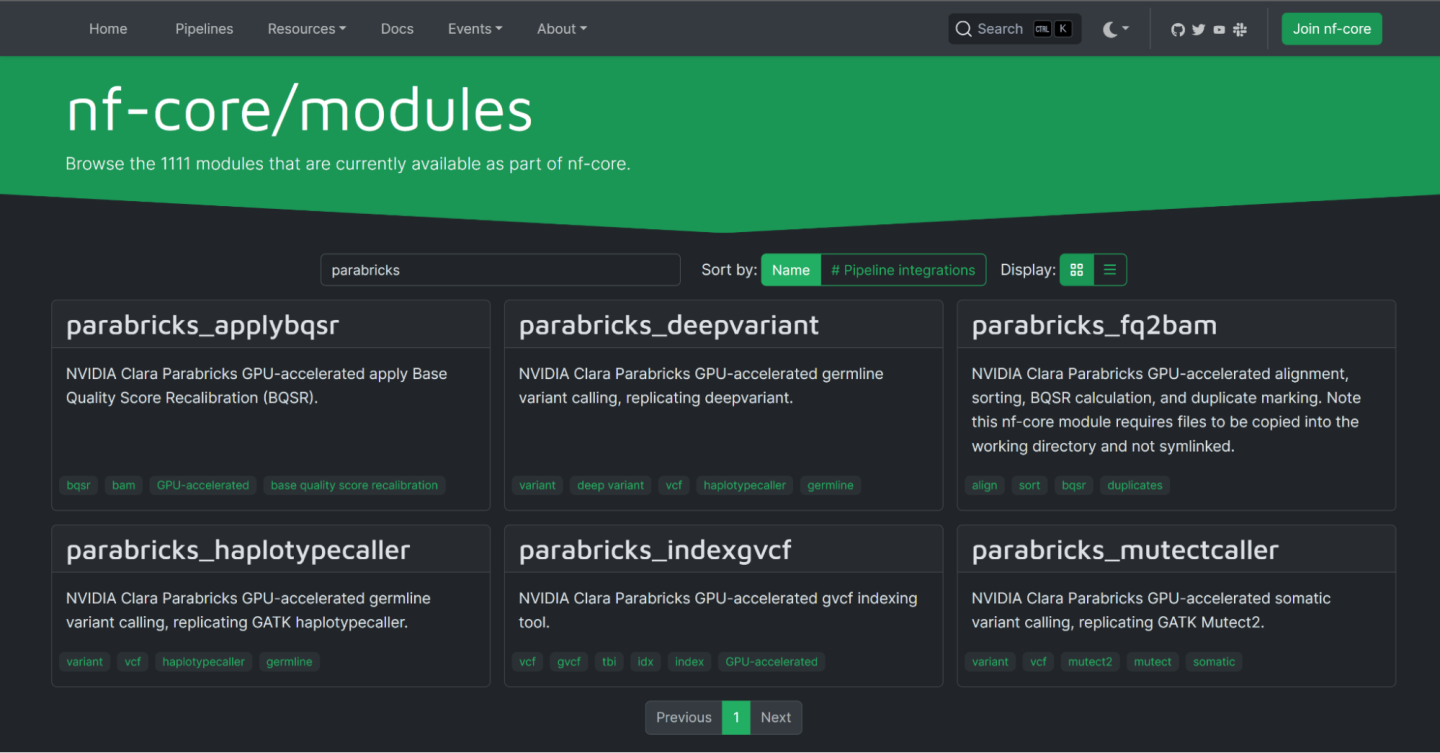
Clicking on any module will take you to the homepage for that module. This page shows the nf-core CLI command to install this module as well as information about the expected inputs and outputs.
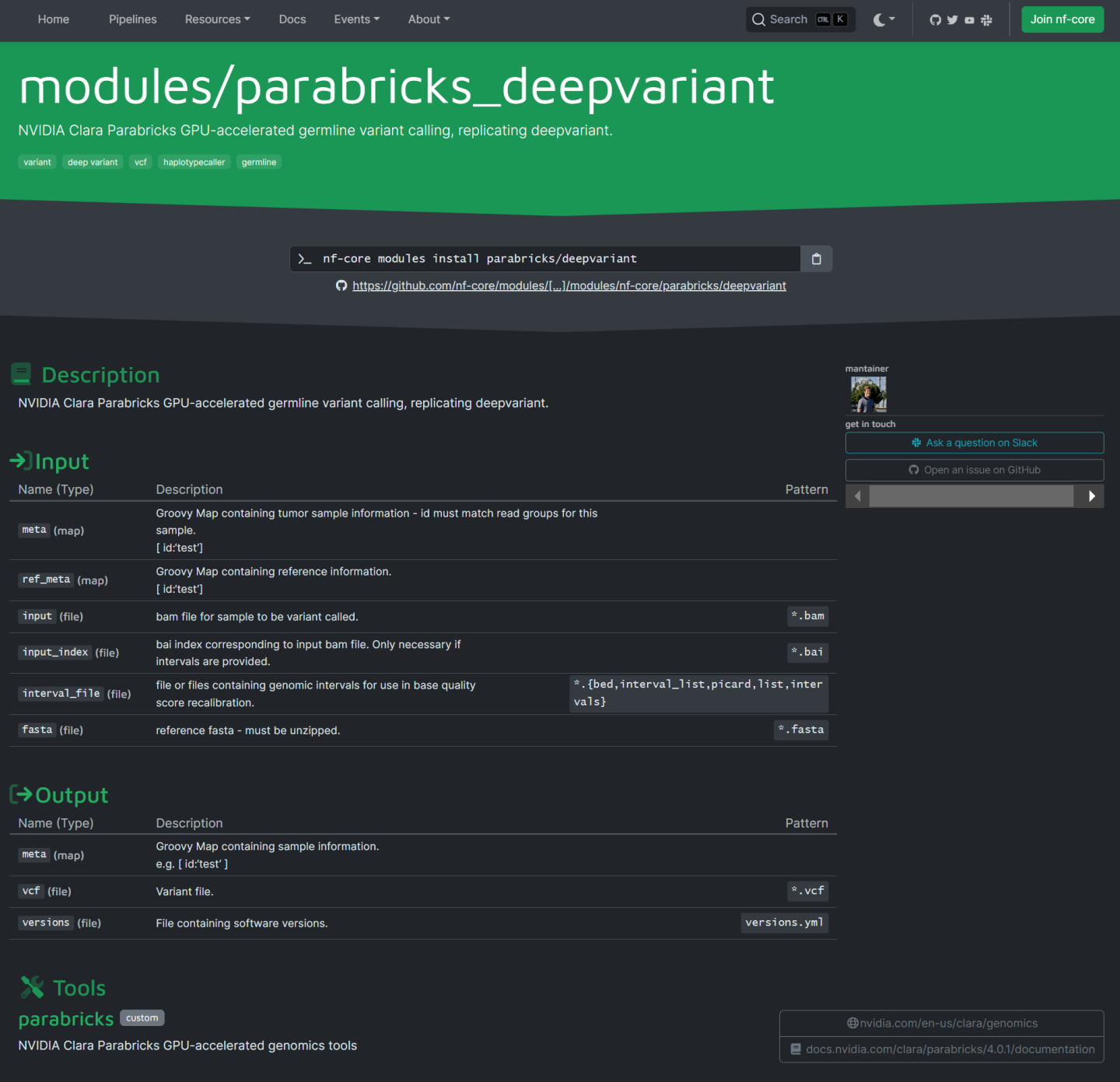
This module can be installed by running:
nf-core modules install parabricks/deepvariant
And it can be tested locally using:
nf-core modules test parabricks/deepvariant
For more details on how to integrate this module into an existing pipeline, see the nf-core documentation.