Denovo PIPELINE
Run the de novo mutation pipeline with 3 samples for de novo variant detection
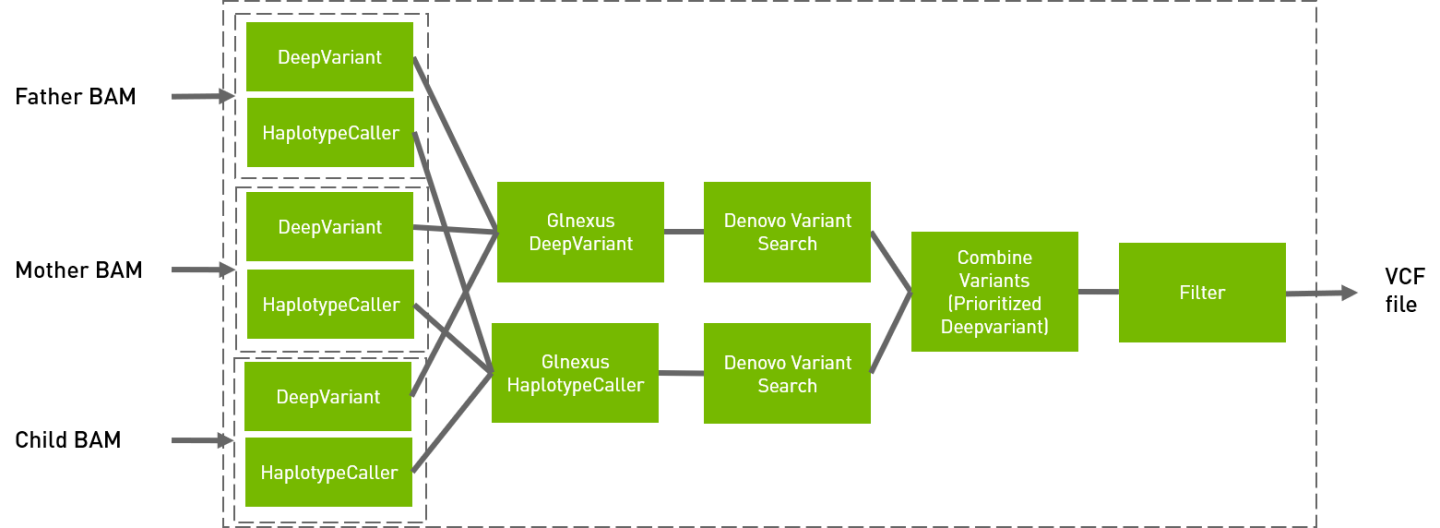
CLI
# The commandline below will run Denovo pipeline.
$ pbrun denovomutation --ref Ref/Homo_sapiens_assembly38.fasta \
--in-mother-bam mother.bam \
--in-father-bam father.bam \
--in-child-bam child.bam \
--out-prefix output
- --ref
- --in-bams
- --in-mother-bam
- --in-father-bam
- --in-child-bam
- --out-prefix
- --disable-deepvariant
- --disable-haplotypecaller
- --pb-model-file
- --disable-use-window-selector-model
- --haplotypecaller-options
- --static-quantized-quals
- --disable-read-filter
- --max-alternate-alleles
- -G, --annotation-group
- -GQB, --gvcf-gq-bands
- --rna
- --dont-use-soft-clipped-bases
- --ploidy
- --dont-use-soft-clipped-bases
- --ploidy PLOIDY
Path to the reference file (default: None)
Paths to the three input BAM/CRAM files for variant calling. Order should be father, mother, and child. Each input file will be run through haplotypecaller and deepvariant (default: None)
Paths to the input BAM/CRAM files for mother (default: None)
Paths to the input BAM/CRAM files for father (default: None)
Paths to the input BAM/CRAM files for child (default: None)
Prefix filename for output data (default: None)
Generate results for haplotypecaller only (default: None)
Generate results for deepvariant only (default: None)
Path of non-default parabricks model file for deepvariant (default: None)
Change the window selector model from Allele Count Linear to Variant Reads. This option will increase the accuracy and runtime (default: None)
Pass supported haplotype caller options as one string. Currently supported original haplotypecaller options: -min-pruning <int>, -standard-min-confidence-threshold-for-calling <int>, -max-reads-per-alignment-start <int>, -min-dangling-branch-length <int>, -pcr-indel-model <NONE, HOSTILE, AGGRESSIVE, CONSERVATIVE>. e.g. –haplotypecaller-options=”-min-pruning 4 -standard-min-confidence-threshold-for-calling 30” (default: None)
Use static quantized quality scores to a given number of levels. Repeat this option multiple times for multiple bins (default: None)
Disable the read filters for bam entries. Currently supported read filters that can be disabled: MappingQualityAvailableReadFilter, MappingQualityReadFilter, NotSecondaryAlignmentReadFilter, WellformedReadFilter (default: None)
Maximum number of alternate alleles to genotype (default: None)
Which groups of annotations to add to the output variant calls. Currently supported annotation groups: StandardAnnotation, StandardHCAnnotation, AS_StandardAnnotation (default: None)
Exclusive upper bounds for reference confidence GQ bands. Must be in the range [1, 100] and specified in increasing order (default: None)
Run haplotypecaller optimized for RNA Data (default: None)
Dont use soft clipped bases for variant calling (default: None)
Ploidy assumed for the bam file. Currently only haploid (ploidy 1) and diploid (ploidy 2) are supported (default: 2)
Dont use soft clipped bases for variant calling.
Ploidy assumed for the bam file. Currently only haploid (ploidy 1) and diploid (ploidy 2) are supported (default: 2)
- --num-gpus NUM_GPUS
- --gpu-devices GPU_DEVICES
Number of GPUs to use for a run. GPUs 0..(NUM_GPUS-1) will be used. If you are using flexera, please include –gpu-devices too.
Which GPU devices to use for a run. By default, all GPU devices will be used. To use specific GPU devices enter a comma-separated list of GPU device numbers. Possible device numbers can be found by examining the output of the nvidia-smi command. For example, using –gpu-devices 0,1 would only use the first two GPUs.
- --tmp-dir TMP_DIR
- --seccomp-override
- --with-petagene-dir WITH_PETAGENE_DIR
- --keep-tmp
- --license-file LICENSE_FILE
- --version
Full path to the directory where temporary files will be stored.
Do not override seccomp options for docker
Full path to the PetaGene installation directory where bin/ and species/ folders are located.
Do not delete the directory storing temporary files after completion.
Path to license file license.bin if not in installation directory.
View compatible software versions.