Decode source and partial (ROI) decoding#
In previous examples we have shown how to decode/read images by using the nvimgcodec.Decoder.decode and nvimgcodec.Decoder.read APIs, to decode from an in-memory encoded stream or a file path, respectively.
[25]:
import os
import numpy as np
from matplotlib import pyplot as plt
resources_dir = os.getenv("PYNVIMGCODEC_EXAMPLES_RESOURCES_DIR", "../../resources/")
img_full_path = os.path.abspath(os.path.join(resources_dir, "jpeg2k/cat-1046544_640.jp2"))
from nvidia import nvimgcodec
decoder = nvimgcodec.Decoder()
def display(images, titles=None, figsize=(10, 4)):
num_images = len(images)
fig, axes = plt.subplots(1, num_images, figsize=figsize)
if num_images == 1:
axes = [axes]
for i, ax in enumerate(axes):
ax.imshow(images[i])
if titles is not None and i < len(titles):
ax.set_title(titles[i])
ax.axis('off')
plt.tight_layout()
plt.show()
We can use the read API to read and decode from a file path:
[26]:
nv_img1 = decoder.read(img_full_path)
Or the decode API to decode from an in-memory encoded stream:
[27]:
with open(img_full_path, 'rb') as in_file:
data = in_file.read()
data = nvimgcodec.DecodeSource(data)
[28]:
nv_img2 = decoder.decode(data)
[29]:
display([nv_img1.cpu(), nv_img2.cpu()], ["Decoder.read", "Decoder.decode"])

These are the simplest methods for interacting with the decoder APIs, which suffice when decoding the entire image. However, there are instances where partial decoding, or region-of-interest (ROI) decoding, is necessary. This approach is particularly crucial for handling very large images, such as satellite, aerial, or medical images, which cannot fit into memory or are impractical to process in their entirety.
nvImageCodec enables users to specify an ROI for the decode API via a generalized nvimgcodec.DecodeSource object. The decode source object includes an encoded stream, which can be an in-memory stream or a file location, and an optional region of interest. Below are examples of using the decode source object without specifying a region of interest:
[30]:
# Decode source is a file path
s1 = nvimgcodec.DecodeSource(img_full_path)
# Decode source is a `bytes` instance (in-memory)
read_bytes = open(img_full_path, 'rb').read()
s2 = nvimgcodec.DecodeSource(read_bytes)
# Decode source is a numpy array (in-memory)
np_arr = np.fromfile(img_full_path, dtype=np.uint8)
s3 = nvimgcodec.DecodeSource(np_arr)
images = [decoder.decode(s).cpu() for s in [s1, s2, s3]]
titles = ['file path', 'bytes', 'np.array']
display(images, titles)

We can now show how to set a region of interest to the decode source location. For the sake of the example, let us find the dimensions of the image, so that we can produce a valid ROI.
[31]:
cs = nvimgcodec.CodeStream(img_full_path)
print(cs.height, 'x', cs.width)
475 x 640
As an example, let us decode a window centered in the image:
[32]:
roi1 = nvimgcodec.Region(int(cs.height * 0.25), int(cs.width * 0.25), int(cs.height * 0.75), int(cs.width * 0.75))
roi2 = nvimgcodec.Region(int(cs.height * 0.2), int(cs.width * 0.2), int(cs.height * 0.5), int(cs.width * 0.5))
decode_sources = [nvimgcodec.DecodeSource(cs, roi) for roi in [roi1, roi2]]
images = [image.cpu() for image in decoder.decode(decode_sources)]
display(images, [str(roi1), str(roi2)])

Bonus: Exploiting tile geometry#
Certain image formats, like TIFF and JPEG2000, allow images to be divided into separate tiles. When requesting an ROI, the alignment between the ROI and these tiles can lead to decoding one or more tiles of the image. In this example, we will demonstrate how to query the image’s tile geometry to align the ROI reading pattern with the image tile boundaries.
[33]:
img_full_path = os.path.abspath(os.path.join(resources_dir, "jpeg2k/tiled-cat-1046544_640.jp2"))
cs = nvimgcodec.CodeStream(img_full_path)
print('Image size:', cs.height, 'x', cs.width)
print('Tile size:', cs.tile_height, 'x', cs.tile_width)
Image size: 475 x 640
Tile size: 100 x 100
When processing a given ROI, the decoder must load and decode the corresponding range of tiles. If the ROI boundary does not align with the tile boundary, the entire tile will be decoded, leading to partial data being discarded.
In scenarios where we can determine the access pattern, we may opt to align the tiled reading with the image’s tile geometry, thereby optimizing resource usage. The example below demonstrates how to create a grid of ROIs that match the image’s tiles.
[34]:
decode_srcs = []
nytiles = (cs.height + cs.tile_height - 1) // cs.tile_height
nxtiles = (cs.width + cs.tile_width - 1) // cs.tile_width
for i in range(nytiles):
row = []
for j in range(nxtiles):
start_y = i * cs.tile_height
end_y = min([(i + 1) * cs.tile_height, cs.height])
start_x = j * cs.tile_width
end_x = min([(j + 1) * cs.tile_width, cs.width])
row.append(
nvimgcodec.DecodeSource(cs, nvimgcodec.Region(start_y, start_x, end_y, end_x))
)
decode_srcs.append(row)
[35]:
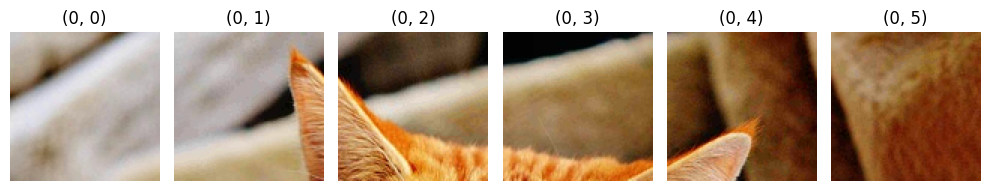
imgs = [img.cpu() for img in decoder.decode(decode_srcs[0][:-1])]
display(imgs, [f"(0, {i})" for i, _ in enumerate(imgs)])
[36]:
imgs = [img.cpu() for img in decoder.decode(decode_srcs[1][:-1])]
display(imgs, [f"(1, {i})" for i, _ in enumerate(imgs)])
