BioNeMo_MegaMolBART Framework
The BioNeMo_MegaMolBART’s training stack is built using CUDA-X, PyTorch, PyTorch lightning, and Apex (for performing training at scale), and NVIDIA’s NeMo framework for building and fine-tuning Large Language models.
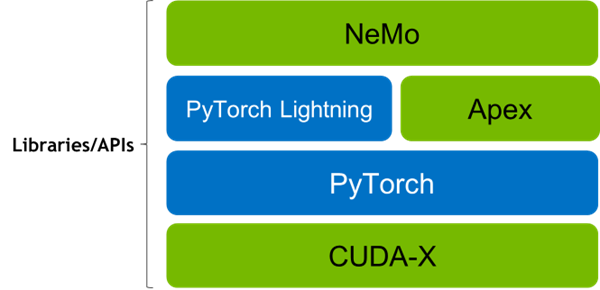
This framework allows distributed model training on a multi-GPU and multi-node compute architecture in a Model parallel, Pipeline parallel, and Data parallel configurations. Setting the desired training configuration is simple as BioNeMo_MegaMolBART uses configurable YAML files as shown below:

Additionally, this framework allows for model checkpointing while training, so users can continue training with a previously trained (or even the provided pre-trained) model.
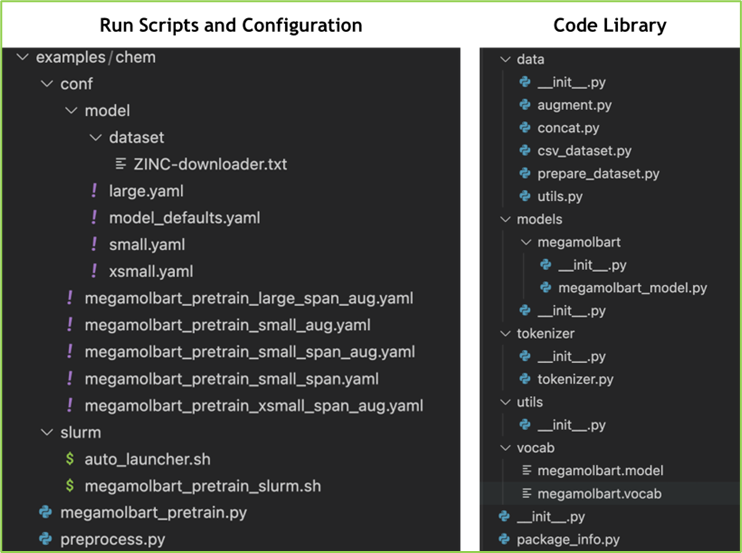
The library organization has the following key components:
examples/chem: It contains configuration files and training scripts
data: It includes classes and functions for loading and augmenting datasets
models: The NeMo MegaMolBART model
tokenizer: MegaMolBART tokenizer for processing SMILES input
vocab: Default vocabulary file and regular expression for tokenizer
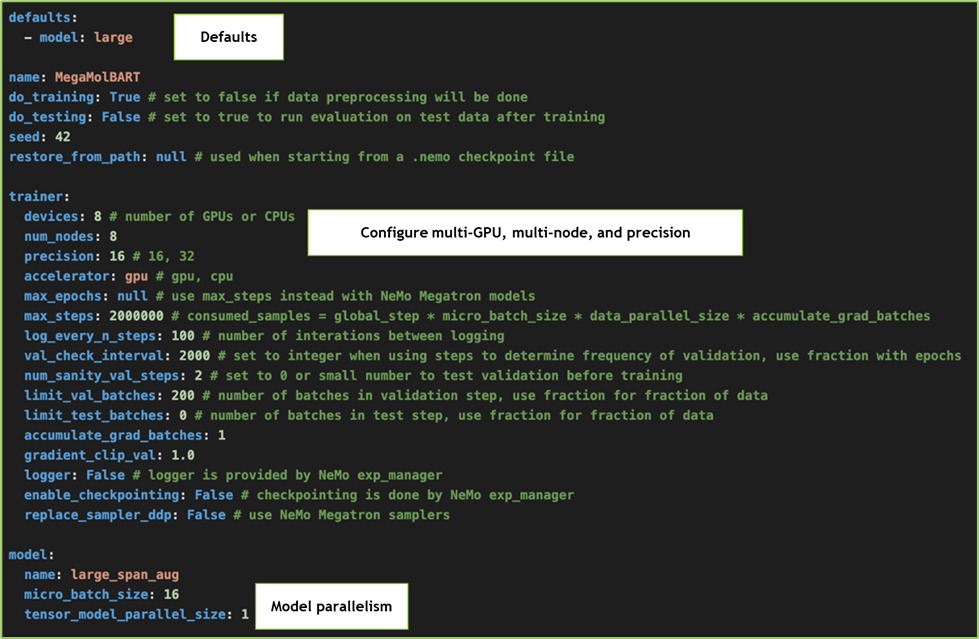
The model configuration file (here, megamolbart_pretrain_large_span_aug.yaml) is in a hierarchical YAML format, as shown in the image. Users can set parameters like devices, nodes, precision levels, etc.
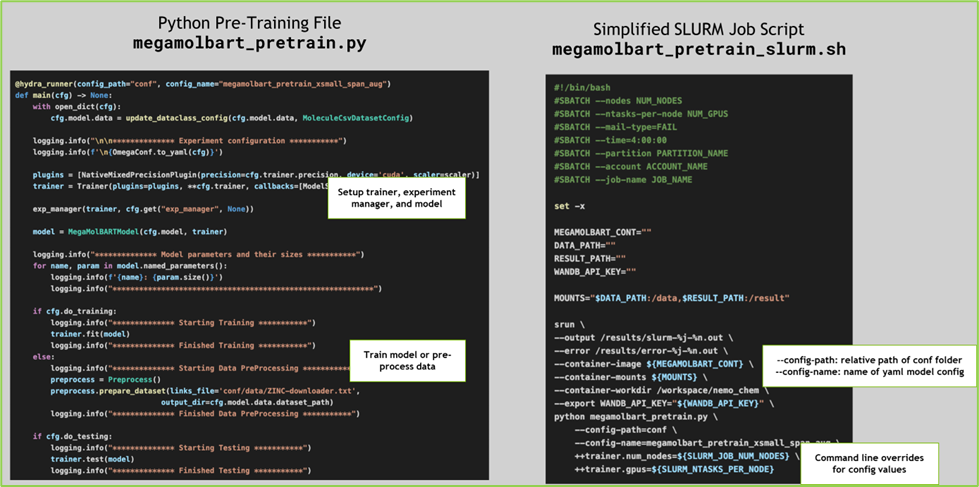
Similarly, the pre-training run script megamolbart_pretrain.py ``in Python; also, a set of scripts for Slurm and Shell (for example, ``megamolbart_pretrain_slurm.sh) are also provided to launch training jobs in respective settings.