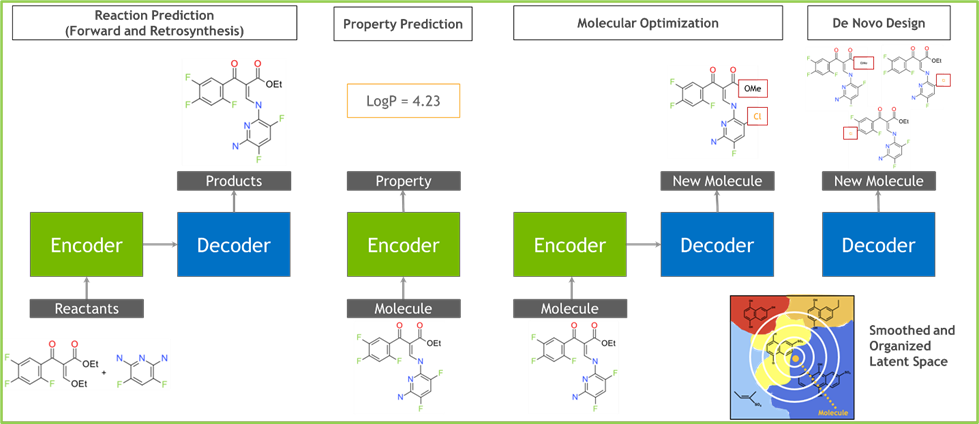
Step #2: Molecule generation and Inference
This section will use the pre-trained BioNeMo_MegaMolBART model to generate new small molecules. In this generative task, which uses both the encoder and the decoder from the pre-trained model, we will obtain the embeddings for the input query SMILES. Once the embeddings are obtained, we will use them to generate analogs/related designs of small molecules for chemical space exploration.
The embedding obtained from the pre-trained model can also be used for modeling and prediction tasks, such as predicting the physicochemical properties of the compounds by using embeddings as input feature sets. The example will walk through the steps – obtaining embeddings, making a prediction model, and model performance comparison with that developed using Morgan fingerprints as the feature set.
To launch the model training on a single node interactively, please go to the Jupyter-Lab link on the left panel:

This will open a new tab in the browser. Click on the directory sign on the left side panel, and then open the “Inference_LP.ipynb”. This will launch the Jupyter notebook in a tab.
Follow the instructions in the Inference_LP.ipynb, and execute each cell by clicking on the cell followed by SHIFT+ENTER.