Clara Train FAQ
Here is a list of frequently asked questions. For more questions including the ability to ask your own, see the NVIDIA Developer Forums:
Researchers can go back to being a data scientist and focus on the problem and not coding
NVIDIA’s Clara Train SDK is optimized as it is written by experienced software developers.
Having reproducible science by sharing simple configuration files
Training speed ups
Simple to deploy in a clinical setting by easily exporting model and importing them into Clara Deploy export tools
Yes, handlers are a list in Clara 4.0, and they will run one by one and may have some dependencies. Please make sure the “write” handler is before the “read” handler according to your training loop.
For example, ValidationHandler should be before CheckpointSaver, otherwise, the best validation metric model cannot be obtained. StatsHandler and TensorBoardHandler are at the end because they only read and print. One note is that “CheckpointSaver” should be before “StatsHandler” if you set “save_final=True” to save model. When an exception happens, ignite only triggers one handler and exits, ignite will add more support in their next version.
Determinism can be set with a random seed. However, if you set “mode=trilinear” in AHNet, determinism is not supported. See PyTorch docs for more details: https://pytorch.org/docs/stable/nn.functional.html#torch.nn.functional.interpolate
Other limitations to determinism may exist, for example the SegResNet implementation in MONAI defaults to upsample_mode: “nontrainable”, which cannot guarantee the model’s reproducibility: https://github.com/Project-MONAI/MONAI/blob/master/monai/networks/nets/segresnet.py#L45
In this case, using upsample_mode: “deconv” can allow for determinism to work.
You can set “factor” on the arg “scale” in your dataset in the configuration to do that:
If factor < 1.0, randomly pick part of the datalist and set to Dataset, useful to quickly test the program.
If factor > 1.0, repeat the datalist to enhance the Dataset.
Example of the usage in MMAR:
"dataset": {
"name": "CacheDataset",
"data_list_file_path": "{DATASET_JSON}",
"data_file_base_dir": "{DATA_ROOT}",
"data_list_key": "training",
"scale": {
"factor": 3.5,
"random_pick": true,
"seed": 123
},
"args": {
"cache_num": 32,
"cache_rate": 0.0,
"num_workers": 4
}
}
Clara 4.0 supports 2 modes in sampling: “auto” - automatically compute weights based on the label count, and “element” - read the weight data from dataset.json directly. It will leverage the PyTorch WeightedRandomSampler to balance the dataset:
"dataset": {
"name": "Dataset",
"data_list_file_path": "{DATASET_JSON}",
"data_file_base_dir": "{DATA_ROOT}",
"data_list_key": "training",
"sampling": {
"mode": "auto",
"num_samples": 50000, # optional, default to the length of dataset on every rank
"replacement": true, # optional, default to True
"label_key": "label", # optional, default to "label"
"weight_key": "weight" # optional, default to "weight"
}
}
For more details, please check: https://github.com/Project-MONAI/MONAI/blob/master/monai/data/samplers.py#L60
If you want to use validation metrics as the indicator for LR ReduceLROnPlateau scheduler, put the “LrScheduleHandler” handler in the “validate” section of config_train.json:
{
"name": "LrScheduleHandler",
"args": {
"print_lr": true,
"epoch_level": true,
"step_transform": "lambda x: x.state.metrics['val_mean_dice']"
}
}
There is a utility in MONAI to support setting different learning rates for different network layers: https://github.com/Project-MONAI/MONAI/blob/master/monai/optimizers/utils.py#L21. To use it in the MMAR config, you can refer to the following as an example:
"optimizer": {
"name": "Adam",
"layer_lrs": [
{
"layer_match": "lambda x: x.model[-1]",
"match_type": "select",
"lr": 1e-3
},
{
"layer_match": "lambda x: 'conv.weight' in x",
"match_type": "filter",
"lr": 5e-4
}
],
"include_others": true,
"args": {
"lr": 1e-4
}
},
TF32 is enabled by default on Ampere GPUs as long as it is not explicitly configured to false. There is a variable and it can be set in the MMAR config: “tf32”: true
Note that with TF32 on, running inference with a checkpoint model may result in minor differences compared to running inference using a torchscript model.
For configuration, set in the MMAR config: “amp”: true
For more details on setting MMAR configs, see Model training and validation configurations. For more details on automatic mixed precision, see: https://developer.nvidia.com/automatic-mixed-precision
If you set “mode=trilinear” in AHNet, AMP=True will be slower than AMP=False on A100 and V100, more details: https://github.com/Project-MONAI/MONAI/issues/1023
Docker comes with dlprof tool see details at https://docs.nvidia.com/deeplearning/frameworks/dlprof-user-guide/#profiling.
Run dlprof train.sh:
You will get a set of files, open nsys_profile.qdrep using the Nsight Systems GUI using vnc (outside your docker)
During training or finetuning, users can set the expected checkpoints to save in the CheckpointSaver handler of the trainer (not evaluator), for example:
{
"name": "CheckpointSaver",
"rank": 0,
"args": {
"save_dir": "{MMAR_CKPT_DIR}",
"save_dict": ["model", "optimizer", "lr_scheduler"],
"save_final": true,
"save_interval": 400
}
}
And during finetuning, users can also load checkpoints for “models” or “optimizer” or “lr_scheduler”:
{
"name": "CheckpointLoader",
"args": {
"load_path": "{MMAR_CKPT}",
"load_dict": ["model", "optimizer"]
}
}
During evaluation, we do not have optimizer and lr_scheduler, and only “model” is available, so since we already set the “CheckpointLoader” handler, there is no need to set it in args again:
{
"name": "CheckpointLoader",
"args": {
"load_path": "{MMAR_CKPT}"
}
}
Yes, the LoadImageD transform can load DICOM images or serials, but it is an experimental feature. You are welcome
to try it and provide feedback.
InvertibleTransform is a highlighted feature of MONAI v0.5, and almost every spatial transform and pad/crop transform can now support inverse operations, including random rotate, random flip, random crop/pad, resize, spacing, orientation, etc.
Users just need to set a “TransformInverter” handler to extract transform context information from specified input data (image or label) field and apply inverse transforms on the expected data (pred or label, etc.):
{
"name": "TransformInverter",
"args": {
"output_keys": ["pred", "label", "pred_class1", "label_class1"],
"batch_keys": "label",
"postfix": "inverted",
"nearest_interp": true
}
}
Then specify the inverted data key when saving segmentation results to files or computing metrics:
{
"name": "MeanDice",
"log_label": "val_mean_dice",
"is_key_metric": true,
"args": {
"include_background": false,
"output_transform": "lambda x: (x['pred_inverted'], x['label_inverted'])"
}
}
{
"name": "SegmentationSaver",
"args": {
"output_dir": "{MMAR_EVAL_OUTPUT_PATH}",
"batch_transform": "lambda x: x['image_meta_dict']",
"output_transform": "lambda x: x['pred_inverted']",
"resample": false
}
}
There is a variable for this feature that can be set in the MMAR config, set: “cudnn_benchmark”: true.
For more information on this variable, see: https://discuss.pytorch.org/t/what-does-torch-backends-cudnn-benchmark-do/5936
For more information on accelerating training, see this PyTorch performance tuning guide.
There is a variable for this feature that can be set in the MMAR config, set: “ddp_find_unused_params”: true.
Please note that this is required to be true for DynUNet, DiNTS, and some other networks for multi-GPU training, but that setting will slow down the model training for most networks in general. For more information about distributed data parallel, see: https://pytorch.org/docs/stable/notes/ddp.html
According to the PyTorch tutorial, users can sync batch normalization during training: https://pytorch.org/docs/stable/generated/torch.nn.SyncBatchNorm.html#torch.nn.SyncBatchNorm.convert_sync_batchnorm
Clara users can achieve this by setting the config variable: “sync_batchnorm”: true.
Yes, you can set a list of losses, a list of models, and a list of optimizers for debug purposes, but you can only enable one in the list, so others should have “disabled=true” set.
If you want to a save final model, please set “save_final=True” for the CheckpointSaver of trainer, not evaluator. The evaluator just works during validation time, it depends on how you config validation intervals.
To avoid repeating the model config in config_validation.json, users can also save the “config_train.json” content into “model.pt” and load the model config from it during valuation.
Just put the “train_conf” key in “CheckpointSaver” of train and validate in config_train.json:
{
"name": "CheckpointSaver",
"rank": 0,
"args": {
"save_dir": "{MMAR_CKPT_DIR}",
"save_dict": ["model", "train_conf"],
"save_key_metric": true
}
}
Then put the path of checkpoint in the model config of config_validation.json:
"model": [
{
"ts_path": "{MMAR_TORCHSCRIPT}",
"disabled": "{dont_load_ts_model}"
},
{
"ckpt_path": "{MMAR_CKPT}",
"disabled": "{dont_load_ckpt_model}"
}
]
Clara 4.0 can achieve EarlyStop based on the “EarlyStopHandler” handler to stop training once a result is good enough.
To use loss value to execute EarlyStop, add the handler to the “train” of config_train.json (added “-” negative value because smaller loss is better):
{
"name": "EarlyStopHandler",
"args": {
"patience": 100,
"score_function": "lambda x: -x.state.output['loss']",
"epoch_level": false
}
}
To use validation metrics (for example: MeanDice) to execute EarlyStop, add the handler to the “validate” section of config_train.json:
{
"name": "EarlyStopHandler",
"args": {
"patience": 1,
"score_function": "lambda x: x.state.metrics['val_mean_dice']"
}
}
For example, the Prostate dataset has 3 output classes, and in order to compute MeanDice on all 3 classes, just add the “SplitChannelsD” transform in the post transform list, referring to the Brats MMAR for more details.
There is a parameter “rank” in handlers of MMAR, set to expected rank index. Please make sure you understand how the handler works, and whether it should run on every rank.
For example:
{
"name": "TensorBoardStatsHandler",
"rank": 0,
"args": {
"log_dir": "{MMAR_CKPT_DIR}",
"tag_name": "train_loss",
"output_transform": "lambda x: x['loss']"
}
}
There are 2 args in SupervisedTrainer and SupervisedEvaluator: “event_names” and “event_to_attr”. User can register their own event names with the “event_names” arg:
"trainer": {
"name": "SupervisedTrainer",
"args": {
"max_epochs": "{epochs}",
"event_names": ["foo_event", "bar_event"]
}
}
Then attach new handler logic to these events and trigger the events at some time, for example here we attached the “test1()” and “test2()” functions to the events and trigger these 2 custom events on the “EPOCH_COMPLETED” event:
class TestHandler:
def attach(self, engine):
engine.add_event_handler("foo_event", self.test1)
engine.add_event_handler("gar_event", self.test2)
engine.add_event_handler(Events.EPOCH_COMPLETED, self)
def __call__(self, engine):
engine.fire_event("foo_event")
engine.fire_event("gar_event")
def test1(self, engine):
pass
def test2(self, engine):
pass
The other arg “event_to_attr” can help register new attributes to “engine.state” associated with the event names. For example, here we register 2 attributes and use them in handlers:
"trainer": {
"name": "SupervisedTrainer",
"args": {
"max_epochs": "{epochs}",
"event_names": ["foo_event", "gar_event"],
"event_to_attr": {"foo_event": "foo", "gar_event": "gar"}
}
}
class TestHandler:
def attach(self, engine):
engine.add_event_handler("foo_event", self.test1)
engine.add_event_handler("gar_event", self.test2)
engine.add_event_handler(Events.EPOCH_COMPLETED, self)
def __call__(self, engine):
engine.fire_event("foo_event")
engine.fire_event("gar_event")
def test1(self, engine):
print(engine.state.foo)
def test2(self, engine):
print(engine.state.gar)
If the transfer learning task has different input or output data classes, we can skip the network layers that have incompatible shape when loading the model checkpoint:
{
"name": "CheckpointLoader",
"args": {
"load_path": "{MMAR_CKPT}",
"load_dict": ["model"],
"strict": false,
"strict_shape": false
}
}
During evaluation, users usually save the metrics of every input image, then analyze the bad cases to improve the deep learning pipeline. To save detailed information of metrics, MONAI provided a handler “MetricsSaver”, which can save the final metric values, raw metric of every model output channel of every input image, metrics summary report of operations: mean, median, max, min, 90percent, std, etc. The MeanDice reports of validation with prostate dataset are as below:
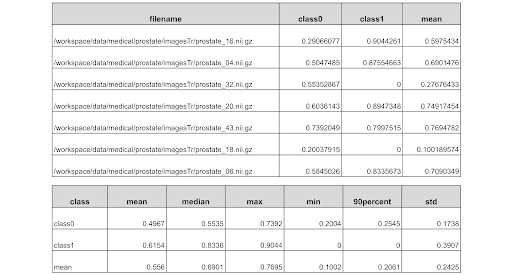
Just set the handler in “config_validation.json”:
{
"name": "MetricsSaver",
"args": {
"save_dir": "{MMAR_EVAL_OUTPUT_PATH}",
"metrics": ["val_mean_dice", "val_acc"],
"metric_details": ["val_mean_dice"],
"batch_transform": "lambda x: x['image_meta_dict']",
"summary_ops": "*",
"save_rank": 0
}
}