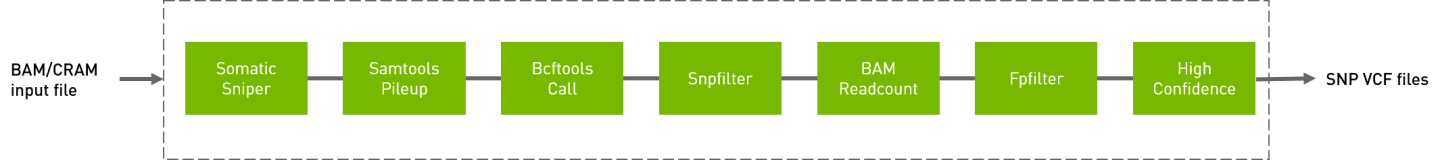
somaticsniper_workflow
Somatic sniper workflow to generate VCF from BAM/CRAM input files.
$ pbrun somaticsniper_workflow \
--ref Ref/Homo_sapiens_assembly38.fasta \
--in-tumor-bam tumor.bam \
--in-normal-bam normal.bam \
--out-prefix output
The commands below are the CPU counterpart of the Parabricks command above. The output from these commands will be identical to the output from the above command. See the Output Comparison page for comparing the results.
bam-somaticsniper -q 1 -G -L -F vcf -f Ref/Homo_sapiens_assembly38.fasta tumor.bam normal.bam output.vcf
bcftools mpileup -A -B -d 2147483647 -Ou -f Ref/Homo_sapiens_assembly38.fasta tumor.bam | bcftools call -c | vcfutils.pl varFilter -Q 20 | awk 'NR > 55 {print}' > output.indel_pileup_Tum.pileup
perl snpfilter.pl --snp-file output.vcf --indel-file output.indel_pileup_Tum.pileup
perl prepare_for_readcount.pl --snp-file output.vcf.SNPfilter
bam-readcount -b 15 -f Ref/Homo_sapiens_assembly38.fasta -l output.vcf.SNPfilter.pos tumor.bam > output.readcounts.rc
perl fpfilter.pl -snp-file output.vcf.SNPfilter -readcount-file output.readcounts.rc
perl highconfidence.pl -snp-file output.vcf.SNPfilter.fp_pass.vcf
Run the somaticsniper variant caller workflow.
Input/Output file options
- --ref REF
- --in-tumor-bam IN_TUMOR_BAM
- --in-normal-bam IN_NORMAL_BAM
- --out-prefix OUT_PREFIX
Path to the reference file. (default: None)
Option is required.
Path of the BAM file for tumor reads. (default: None)
Option is required.
Path of the BAM file for normal reads. (default: None)
Option is required.
Prefix filename for output data. (default: None)
Option is required.
Tool Options:
- --num-threads NUM_THREADS
- --min-mapq MIN_MAPQ
Number of threads for worker. (default: 1)
Filter reads with mapping quality less than this value. (default: 1)
Common options:
- --logfile LOGFILE
- --tmp-dir TMP_DIR
- --with-petagene-dir WITH_PETAGENE_DIR
- --keep-tmp
- --license-file LICENSE_FILE
- --no-seccomp-override
- --version
Path to the log file. If not specified, messages will only be written to the standard error output. (default: None)
Full path to the directory where temporary files will be stored.
Full path to the PetaGene installation directory. By default, this should have been installed at /opt/petagene. Use of this option also requires that the PetaLink library has been preloaded by setting the LD_PRELOAD environment variable. Optionally set the PETASUITE_REFPATH and PGCLOUD_CREDPATH environment variables that are used for data and credentials (default: None)
Do not delete the directory storing temporary files after completion.
Path to license file license.bin if not in the installation directory.
Do not override seccomp options for docker (default: None).
View compatible software versions.