germline (GATK Germline Pipeline)
GATK, the Genome Analysis Toolkit, is an industry standard software package developed by the Broad Institute of MIT and Harvard and designed to be used for a wide range of genomic analyses, including variant discovery, genotyping, and more. GATK is one of the most popular tools used in bioinformatics for analyzing next-generation sequencing datasets and is an industry standard for calling single nucleotide variants (SNVs) and insertions/deletions (InDels) from sequencing data in germline samples.
GATK offers robust, accurate analysis of sequencing data and is frequently updated to include the latest best practices for variant discovery. With high reliability and the ability to be used for a number of use cases, GATK is a gold standard tool for any researcher working with next-generation sequencing data.
The GATK germline workflow for variant calling can be deployed within NVIDIA’s Parabricks software suite, which is designed for accelerated secondary analysis in genomics, bringing industry standard tools and workflows from CPU to GPU, and delivering the same results at up to 60x faster runtimes. A 30x whole genome can be analyzed in under 25 minutes on an NVIDIA DGX system, compared to over 30 hours on a CPU instance (m5.24xlarge, 96 x vCPU), and exomes can be analyzed in just 4 minutes. This means Parabricks running on one NVIDIA DGX A100, can analyze up to 25,000 whole genomes per year. The NVIDIA team collaborated with the GATK team at the Broad Institute to evaluate the accuracy of germline workflows. Through this rigorous process, they verified that the Parabricks workflows produce results that are functionally equivalent to the CPU-native GATK versions.
As a specific example, benchmarking on publicly available Genome in a Bottle (GIAB) samples with the fq2bam and germline caller workflows from the Parabricks suite produced variant calling results that were >0.9999 equivalent in both precision and recall to those produced by the BWA, MarkDuplicates, BQSR, and HaplotypeCaller commands in the GATK’s Whole Genome Germline Single Sample variant calling workflow.
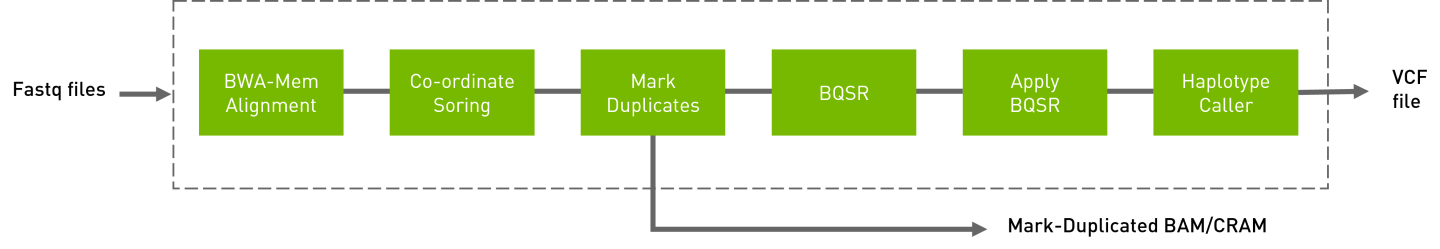
Given one or more pairs of FASTQ files, you can run the germline variant tool to generate BAM, variants, duplicate metrics and recal.
The germline pipeline shown below resembles the GATK4 best practices pipeline. The inputs are BWA-indexed reference files, pair-ended FASTQ files, and knownSites for BQSR calculation. The outputs of this pipeline are as follows:
Aligned, co-ordinate sorted, duplicated marked BAM
BQSR report
Variants in
vcf/g.vcf/g.vcf.gzformat
Running the germline pipeline:
# This command assumes all the inputs are in INPUT_DIR and all the outputs go to OUTPUT_DIR.
$ docker run --rm --gpus all --volume INPUT_DIR:/workdir --volume OUTPUT_DIR:/outputdir \
--workdir /workdir \
nvcr.io/nvidia/clara/clara-parabricks:4.1.0-1 \
pbrun germline \
--ref /workdir/${REFERENCE_FILE} \
--in-fq /workdir/${INPUT_FASTQ_1} /workdir/${INPUT_FASTQ_2} \
--knownSites /workdir/${KNOWN_SITES_FILE} \
--out-bam /outputdir/${OUTPUT_BAM} \
--out-variants /outputdir/${OUTPUT_VCF} \
--out-recal-file /outputdir/${OUT_RECAL_FILE}
Several original HaplotypeCaller options are supported by Parabricks.
To specify the inclusion or exclusion of several haplotype caller annotations,
use the --haplotypecaller-options option:
# This command assumes all the inputs are in INPUT_DIR and all the outputs go to OUTPUT_DIR.
$ docker run --rm --gpus all --volume INPUT_DIR:/workdir --volume OUTPUT_DIR:/outputdir \
--workdir /workdir \
nvcr.io/nvidia/clara/clara-parabricks:4.1.0-1 \
pbrun haplotypecaller \
...
--haplotypecaller-options '-min-pruning 4 -A AS_BaseQualityRankSumTest -A TandemRepeat'
...
Annotations may be excluded in the same manner using the -AX option. There should
be a space between the -A/-AX flag and its value.
The following are supported options and their allowed values:
- -A
AS_BaseQualityRankSumTest
AS_FisherStrand
AS_InbreedingCoeff
AS_MappingQualityRankSumTest
AS_QualByDepth
AS_RMSMappingQuality
AS_ReadPosRankSumTest
AS_StrandOddsRatio
BaseQualityRankSumTest
ChromosomeCounts
ClippingRankSumTest
Coverage
DepthPerAlleleBySample
DepthPerSampleHC
ExcessHet
FisherStrand
InbreedingCoeff
MappingQualityRankSumTest
QualByDepth
RMSMappingQuality
ReadPosRankSumTest
ReferenceBases
StrandBiasBySample
StrandOddsRatio
TandemRepeat
- -AX
(same as for the -A option)
- --output-mode
EMIT_VARIANTS_ONLY
EMIT_ALL_CONFIDENT_SITES
EMIT_ALL_ACTIVE_SITES
- -max-reads-per-alignment-start
a positive integer
- -min-dangling-branch-length
a positive integer
- -min-pruning
a positive integer
- -pcr-indel-model
NONE
HOSTILE
AGGRESSIVE
CONSERVATIVE
- -standard-min-confidence-threshold-for-calling
a positive integer
The commands below are the bwa-0.7.12 and GATK4 counterpart of the Parabricks command above. The output from these commands will be identical to the output from the above command. See the Output Comparison page for comparing the results.
# Run bwa-mem and pipe output to create sorted BAM
$ bwa mem \
-t 32 \
-K 10000000 \
-R '@RG\tID:sample_rg1\tLB:lib1\tPL:bar\tSM:sample\tPU:sample_rg1' \
<INPUT_DIR>/${REFERENCE_FILE} <INPUT_DIR>/${INPUT_FASTQ_1} <INPUT_DIR>/${INPUT_FASTQ_2} | \
gatk SortSam \
--java-options -Xmx30g \
--MAX_RECORDS_IN_RAM 5000000 \
-I /dev/stdin \
-O cpu.bam \
--SORT_ORDER coordinate
# Mark Duplicates
$ gatk MarkDuplicates \
--java-options -Xmx30g \
-I cpu.bam \
-O mark_dups_cpu.bam \
-M metrics.txt
# Generate BQSR Report
$ gatk BaseRecalibrator \
--java-options -Xmx30g \
--input mark_dups_cpu.bam \
--output <OUTPUT_DIR>/${OUT_RECAL_FILE} \
--known-sites <INPUT_DIR>/${KNOWN_SITES_FILE} \
--reference <INPUT_DIR>/${REFERENCE_FILE}
# Run ApplyBQSR Step
$ gatk ApplyBQSR \
--java-options -Xmx30g \
-R <INPUT_DIR>/${REFERENCE_FILE} \
-I mark_dups_cpu.bam \
--bqsr-recal-file <OUTPUT_DIR>/${OUT_RECAL_FILE} \
-O cpu_nodups_BQSR.bam
#Run Haplotype Caller
$ gatk HaplotypeCaller \
--java-options -Xmx30g \
--input cpu_nodups_BQSR.bam \
--output <OUTPUT_DIR>/${OUTPUT_VCF} \
--reference <INPUT_DIR>/${REFERENCE_FILE} \
--native-pair-hmm-threads 16
Run Germline pipeline to convert FASTQ to VCF.
Input/Output file options
- --ref REF
- --in-fq [IN_FQ [IN_FQ ...]]
- --in-se-fq [IN_SE_FQ [IN_SE_FQ ...]]
- --knownSites KNOWNSITES
- --interval-file INTERVAL_FILE
- --out-recal-file OUT_RECAL_FILE
- --out-bam OUT_BAM
- --out-variants OUT_VARIANTS
- --out-duplicate-metrics OUT_DUPLICATE_METRICS
Path to the reference file. (default: None)
Option is required.
Path to the pair-ended FASTQ files followed by optional read groups with quotes (Example: "@RGtID:footLB:lib1tPL:bartSM:sampletPU:foo"). The files must be in fastq or fastq.gz format. All sets of inputs should have a read group; otherwise, none should have a read group, and it will be automatically added by the pipeline. This option can be repeated multiple times. Example 1: --in-fq sampleX_1_1.fastq.gz sampleX_1_2.fastq.gz --in-fq sampleX_2_1.fastq.gz sampleX_2_2.fastq.gz. Example 2: --in-fq sampleX_1_1.fastq.gz sampleX_1_2.fastq.gz "@RGtID:footLB:lib1tPL:bartSM:sampletPU:unit1" --in-fq sampleX_2_1.fastq.gz sampleX_2_2.fastq.gz "@RGtID:foo2tLB:lib1tPL:bartSM:sampletPU:unit2". For the same sample, Read Groups should have the same sample name (SM) and a different ID and PU. (default: None)
Path to the single-ended FASTQ file followed by optional read group with quotes (Example: "@RGtID:footLB:lib1tPL:bartSM:sampletPU:foo"). The file must be in fastq or fastq.gz format. Either all sets of inputs have a read group, or none should have one, and it will be automatically added by the pipeline. This option can be repeated multiple times. Example 1: --in-se-fq sampleX_1.fastq.gz --in-se-fq sampleX_2.fastq.gz . Example 2: --in-se-fq sampleX_1.fastq.gz "@RGtID:footLB:lib1tPL:bartSM:sampletPU:unit1" --in-se-fq sampleX_2.fastq.gz "@RGtID:foo2tLB:lib1tPL:bartSM:sampletPU:unit2" . For the same sample, Read Groups should have the same sample name (SM) and a different ID and PU. (default: None)
Path to a known indels file. The file must be in vcf.gz format. This option can be used multiple times. (default: None)
Path to an interval file in one of these formats: Picard-style (.interval_list or .picard), GATK-style (.list or .intervals), or BED file (.bed). This option can be used multiple times. (default: None)
Path of the report file after Base Quality Score Recalibration. (default: None)
Path of BAM file after Marking Duplicates. (default: None)
Option is required.
Path of the vcf/gvcf/gvcf.gz file after variant calling. (default: None)
Option is required.
Path of duplicate metrics file after Marking Duplicates. (default: None)
Tool Options:
- -L INTERVAL, --interval INTERVAL
- --bwa-options BWA_OPTIONS
- --no-warnings
- --gpuwrite
- --gpusort
- --low-memory
- --filter-flag FILTER_FLAG
- --skip-multiple-hits
- --min-read-length MIN_READ_LENGTH
- --align-only
- --no-markdups
- --fix-mate
- --markdups-assume-sortorder-queryname
- --markdups-picard-version-2182
- --optical-duplicate-pixel-distance OPTICAL_DUPLICATE_PIXEL_DISTANCE
- --read-group-sm READ_GROUP_SM
- --read-group-lb READ_GROUP_LB
- --read-group-pl READ_GROUP_PL
- --read-group-id-prefix READ_GROUP_ID_PREFIX
- -ip INTERVAL_PADDING, --interval-padding INTERVAL_PADDING
- --haplotypecaller-options HAPLOTYPECALLER_OPTIONS
- --static-quantized-quals STATIC_QUANTIZED_QUALS
- --gvcf
- --batch
- --disable-read-filter DISABLE_READ_FILTER
- --max-alternate-alleles MAX_ALTERNATE_ALLELES
- -G ANNOTATION_GROUP, --annotation-group ANNOTATION_GROUP
- -GQB GVCF_GQ_BANDS, --gvcf-gq-bands GVCF_GQ_BANDS
- --rna
- --dont-use-soft-clipped-bases
- --read-from-tmp-dir
- --run-partition
- --no-alt-contigs
- --ploidy PLOIDY
Interval within which to call bqsr from the input reads. All intervals will have a padding of 100 to get read records, and overlapping intervals will be combined. Interval files should be passed using the --interval-file option. This option can be used multiple times (e.g. "-L chr1 -L chr2:10000 -L chr3:20000+ -L chr4:10000-20000"). (default: None)
Pass supported bwa mem options as one string. The current original bwa mem supported options are -M, -Y and -T (e.g. --bwa-options="-M -Y") (default: None)
Suppress warning messages about system thread and memory usage. (default: None)
Use one GPU to accelerate writing final BAM. (default: None)
Use GPUs to accelerate sorting and marking. (default: None)
Use low memory mode. (default: None)
Don't generate SAM entries in the output if the entry's flag's meet this criteria. Criteria: (flag & filter != 0) (default: 0)
Filter SAM entries whose length of SA is not 0. (default: None)
Skip reads below minimum read length. They will not be part of the output. (default: None)
Generate output BAM after bwa-mem. The output will not be co-ordinate sorted or duplicates will not be marked. (default: None)
Do not perform the Mark Duplicates step. Return BAM after sorting. (default: None)
Add mate cigar (MC) and mate quality (MQ) tags to the output file. (default: None)
Assume the reads are sorted by queryname for Marking Duplicates. This will mark secondary, supplementary, and unmapped reads as duplicates as well. This flag will not impact variant calling while increasing processing times. (default: None)
Assume marking duplicates to be similar to Picard version 2.18.2. (default: None)
The maximum offset between two duplicate clusters in order to consider them optical duplicates. Ignored if --out-duplicate-metrics is not passed. (default: None)
SM tag for read groups in this run. (default: None)
LB tag for read groups in this run. (default: None)
PL tag for read groups in this run. (default: None)
Prefix for the ID and PU tags for read groups in this run. This prefix will be used for all pairs of fastq files in this run. The ID and PU tags will consist of this prefix and an identifier, that will be unique for a pair of fastq files. (default: None)
Amount of padding (in base pairs) to add to each interval you are including. (default: None)
Pass supported haplotype caller options as one string. The following are currently supported original haplotypecaller options: -A <AS_BaseQualityRankSumTest, AS_FisherStrand, AS_InbreedingCoeff, AS_MappingQualityRankSumTest, AS_QualByDepth, AS_RMSMappingQuality, AS_ReadPosRankSumTest, AS_StrandOddsRatio, BaseQualityRankSumTest, ChromosomeCounts, ClippingRankSumTest, Coverage, DepthPerAlleleBySample, DepthPerSampleHC, ExcessHet, FisherStrand, InbreedingCoeff, MappingQualityRankSumTest, QualByDepth, RMSMappingQuality, ReadPosRankSumTest, ReferenceBases, StrandBiasBySample, StrandOddsRatio, TandemRepeat>,-AX <same options as -A>,--output-mode <EMIT_VARIANTS_ONLY, EMIT_ALL_CONFIDENT_SITES, EMIT_ALL_ACTIVE_SITES> ,-max-reads-per-alignment-start <int>, -min-dangling-branch-length <int>, -min-pruning <int>, -pcr-indel-model <NONE, HOSTILE, AGGRESSIVE, CONSERVATIVE>, -standard-min-confidence-threshold-for-calling <int>(e.g. --haplotypecaller-options="-min-pruning 4 -standard-min-confidence-threshold-for-calling 30"). (default: None)
Use static quantized quality scores to a given number of levels. Repeat this option multiple times for multiple bins. (default: None)
Generate variant calls in .gvcf Format. (default: None)
Given an input list of BAMs, run the variant calling of each BAM using one GPU, and process BAMs in parallel based on how many GPUs the system has. (default: None)
Disable the read filters for BAM entries. Currently, the supported read filters that can be disabled are MappingQualityAvailableReadFilter, MappingQualityReadFilter, NotSecondaryAlignmentReadFilter, and WellformedReadFilter. (default: None)
Maximum number of alternate alleles to genotype. (default: None)
The groups of annotations to add to the output variant calls. Currently supported annotation groups are StandardAnnotation, StandardHCAnnotation, and AS_StandardAnnotation. (default: None)
Exclusive upper bounds for reference confidence GQ bands. Must be in the range [1, 100] and specified in increasing order. (default: None)
Run haplotypecaller optimized for RNA data. (default: None)
Don't use soft clipped bases for variant calling. (default: None)
Read from the temporary files generated by fq2bam. (default: None)
Divide the whole genome into multiple partitions and run multiple processes at the same time, each on one partition. (default: None)
Get rid of output records for alternate contigs. (default: None)
Ploidy assumed for the BAM file. Currently only haploid (ploidy 1) and diploid (ploidy 2) are supported. (default: 2)
Common options:
- --logfile LOGFILE
- --tmp-dir TMP_DIR
- --with-petagene-dir WITH_PETAGENE_DIR
- --keep-tmp
- --no-seccomp-override
- --version
Path to the log file. If not specified, messages will only be written to the standard error output. (default: None)
Full path to the directory where temporary files will be stored.
Full path to the PetaGene installation directory. By default, this should have been installed at /opt/petagene. Use of this option also requires that the PetaLink library has been preloaded by setting the LD_PRELOAD environment variable. Optionally set the PETASUITE_REFPATH and PGCLOUD_CREDPATH environment variables that are used for data and credentials (default: None)
Do not delete the directory storing temporary files after completion.
Do not override seccomp options for docker (default: None).
View compatible software versions.
GPU options:
- --num-gpus NUM_GPUS
Number of GPUs to use for a run. GPUs 0..(NUM_GPUS-1) will be used.
The --in-fq option takes the names of two FASTQ files, optionally followed by a quoted read group. The FASTQ filenames must not start with a hyphen.
In the values provided to --haplotypecaller-options --output-mode requires two leading hyphens, while all other values take a single hyphen.