Holoscan Sample Applications
This section provides details on the Holoscan sample applications:
Refer to the instructions in the Holoscan NGC container overview or the Github source repository to run the application, depending on how you’ve chosen to set up Holoscan.
Digital endoscopy is a key technology for medical screenings and minimally invasive surgeries. Using real-time AI workflows to process and analyze the video signal produced by the endoscopic camera, this technology helps medical professionals with anomaly detection and measurements, image enhancements, alerts, and analytics.

Fig. 18 Endoscopy image from a gallbladder surgery showing AI-powered frame-by-frame tool identification and tracking. Image courtesy of Research Group Camma, IHU Strasbourg and the University of Strasbourg
The Endoscopy tool tracking application provides an example of how an endoscopy data stream can be captured and processed using the C++ or Python APIs on multiple hardware platforms.

Fig. 19 Tool tracking application workflow with replay from file
The pipeline uses a recorded endoscopy video file (generated by convert_video_to_gxf_entities script) for input frames.
Each input frame in the file is loaded by Video Stream Replayer and Broadcast node passes the frame to the following two nodes (Entities):
Format Converter: Convert image format from
RGB888(24-bit pixel) toRGBA8888(32-bit pixel) for visualization (Tool Tracking Visualizer)Format Converter: Convert the data type of the image from
uint8tofloat32for feeding into the tool tracking model (by Custom TensorRT Inference)
Then, Tool Tracking Visualizer uses outputs from the first Format Converter and Custom TensorRT Inference to render overlay frames (mask/point/text) on top of the original video frames.

Fig. 20 AJA tool tracking app
The pipeline is similar with Input source: Video Stream Replayer but the input source is replaced with AJA Source.
The pipeline graph also defines an optional Video Stream Recorder that can be enabled to record the original video stream to disk. This stream recorder (and its associated Format Converter) are commented out in the graph definition and thus are disabled by default in order to maximize performance. To enable the stream recorder, uncomment all of the associated components in the graph definition.
AJA Source: Get video frame from AJA HDMI capture card (pixel format is
RGBA8888with the resolution of 1920x1080)Format Converter: Convert image format from
RGB8888(32-bit pixel) toRGBA888(24-bit pixel) for recording (Video Stream Recorder)Video Stream Recorder: Record input frames into a file
The Hi-Speed Endoscopy Application is not included in the Holoscan container on NGC. Instead, refer to the run instructions in the Github source repository.
The hi-speed endoscopy application showcases how high resolution cameras can be used to capture the scene, post-processed on GPU, and displayed at high frame rate.
This application requires:
an Emergent Vision Technologies camera (see setup instructions)
an NVIDIA ConnectX SmartNIC with Rivermax SDK and drivers installed
a display with high refresh rate to keep up with the camera’s framerate
additional setups to reduce latency
Tested on the Holoscan DevKits (ConnectX included) with:
EVT HB-9000-G-C: 25GigE camera with Gpixel GMAX2509
SFP28 cable and QSFP28 to SFP28 adaptor
Asus ROG Swift PG279QM and Asus ROG Swift 360 Hz PG259QNR monitors with NVIDIA G-SYNC technology
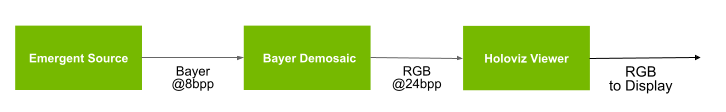
Fig. 21 Hi-Speed Endoscopy App
The data acquisition happens using emergent-source, by default it is set to 4200x2160 at 240Hz.
The acquired data is then demosaiced in GPU using CUDA via bayer-demosaic and displayed through
holoviz-viewer.
The peak performance that can be obtained by running these applications with the recommended hardware, GSYNC and RDMA enabled on exclusive display mode is 10ms on Clara AGX Devkit and 8ms on NVIDIA IGX Orin DevKit ES. This is the latency of a frame from scene acquisition to display on monitor.
Troubleshooting
Problem: The application fails to find the EVT camera.
Make sure that the MLNX ConnectX SmartNIC is configured with the correct IP address. Follow section Post EVT Software Installation Steps
Problem: The application fails to open the EVT camera.
Make sure that the application was run with
sudoprivileges.Make sure a valid Rivermax license file is located at
/opt/mellanox/rivermax/rivermax.lic.
Problem: The application fails to connect to the EVT camera with error message “GVCP ack error”.
It could be an issue with the HR12 power connection to the camera. Disconnect the HR12 power connector from the camera and try reconnecting it.
Refer to the instructions in the Holoscan NGC container overview or the Github source repository to run the application, depending on how you’ve chosen to set up Holoscan.
This section describes the details of the ultrasound segmentation sample application as well as how to load a custom inference model into the application for some limited customization. Out of the box, the ultrasound segmentation application comes as a “video replayer” and “AJA source”, where the user can replay a pre-recorded ultrasound video file included in the holoscan container or stream data from an AJA capture device directly through the GPU respectively.
This application performs an automatic segmentation of the spine from a trained AI model for the purpose of scoliosis visualization and measurement.
This application is available in C++ and Python API variants.

Fig. 22 Spine segmentation of ultrasound data
Input source: Video Stream Replayer

Fig. 23 Segmentation application with replay from file
The pipeline uses a pre-recorded endoscopy video stream stored in nvidia::gxf::Tensor format as input. The tensor-formatted file is generated via convert_video_to_gxf_entities from a pre-recorded MP4 video file.
Input frames are loaded by Video Stream Replayer and Broadcast node passes the frame to two branches in the pipeline.
In the inference branch the video frames are converted to floating-point precision using the format converter, pixel-wise segmentation is performed, and the segmentation result if post-processed for the visualizer.
The visualizer receives the original frame as well as the result of the inference branch to show an overlay.

Fig. 24 AJA segmentation app
This pipeline is exactly the same as the pipeline described in the previous section except the Video Stream Replayer has been substituted with an AJA Video Source.
Bring Your Own Model (BYOM) - Customizing the Ultrasound Segmentation Application For Your Model
This section shows how the user can easily modify the ultrasound segmentation app to run a different segmentation model, even of an entirely different modality. In this use case we will use the ultrasound application to implement a polyp segmentation model to run on a Colonoscopy sample video.
At this time the holoscan containers contain only binaries of the sample applications, meaning users may not modify the extensions. However, the users can substitute the ultrasound model with their own and add, remove, or replace the extensions used in the application.
As a first step, please go to the Colonoscopy Sample Application Data NGC Resource to download the model and video data.
For a comprehensive guide on building your own Holoscan extensions and applications please refer to the Using the SDK section of the guide.
The sample ultrasound segmentation model expects a gray-scale image of 256 x 256 and outputs a semantic segmentation of the same size with two channels representing bone contours with hyperechoic lines (foreground) and hyperechoic acoustic shadow (background).
Currently, the sample apps are able to load ONNX models, or TensorRT engine files built for the architecture on which you will be running the model only. TRT engines are automatically generated from ONNX by the application when it is run.
If you are converting your model from PyTorch to ONNX, chances are your input is NCHW, and will need to be converted to NHWC. An example transformation script is included with the colonoscopy sample downloaded above, and is found inside the resource as model/graph_surgeon.py. You may need to modify the dimensions as needed before modifying your model as:
python graph_surgeon.py {YOUR_MODEL_NAME}.onnx {DESIRED_OUTPUT_NAME}.onnx
Note that this step is optional if you are directly using ONNX models.
To get a better understanding of the model, applications such as netron.app can be used.
We will now substitute the model and sample video to inference upon as follows.
Enter the holoscan container, but make sure to load the colonoscopy model from the host into the container. Assuming your model is in
${my_model_path_dir}and your data is in${my_data_path_dir}then you can execute the following:docker run -it --rm --runtime=nvidia \ -e NVIDIA_DRIVER_CAPABILITIES=graphics,video,compute,utility \ -v ${my_model_path_dir}:/workspace/my_model \ -v ${my_data_path_dir}:/workspace/my_data \ -v /tmp/.X11-unix:/tmp/.X11-unix \ -e DISPLAY=${DISPLAY} \ nvcr.io/nvidia/clara-holoscan/holoscan:v0.4.0
Check that the model and data correctly appear under
/workspace/my_modeland/workspace/my_data.Now we are ready to make the required modifications to the ultrasound sample application to have the colonoscopy model load.
cd /opt/nvidia/holoscan vi ./apps/ultrasound_segmentation/gxf/segmentation_replayer.yaml
In the editor navigate to the first entity
source, and under typenvidia::holoscan::stream_playback::VideoStreamReplayerwe will modify the following for our input video:a.
directory: "/workspace/my_data"b.
basename: "colonoscopy"TipIn general, to be able to play a desired video through a custom model we first need to convert the video file into a GXF replayable tensor format. This step has already been done for the colonoscopy example, but for a custom video perform the following actions inside the container.
apt update && DEBIAN_FRONTEND=noninteractive apt install -y ffmpeg cd /workspace git clone https://github.com/nvidia-holoscan/holoscan-sdk.git cd clara-holoscan-embedded-sdk/scripts ffmpeg -i /workspace/my_data/${my_video} -pix_fmt rgb24 -f rawvideo pipe:1 | python3 convert_video_to_gxf_entities.py --width ${my_width} --height ${my_height} --directory /workspace/my_data --basename my_video
The above commands should yield two Holoscan tensor replayer files in
/workspace/my_data, namelymy_video.gxf_indexandmy_video.gxf_entities.In the editor navigate to the
segmentation_preprocessorentity. Under typenvidia::holoscan::formatconverter::FormatConverterwe will modify the following parameters to fit the input dimensions of our colonoscopy model:a.
resize_width: 512b.
resize_height: 512In the editor navigate to the
segmentation_inferenceentity. We will modify thenvidia::gxf::TensorRtInferencetype where we want to specify the input and output names.a. Specify the location of your
ONNXfiles as:model_file_path: /workspace/my_model/colon.onnxb. Specify the location of TensorRT engines as:
engine_cache_dir: /workspace/my_model/cachec. Specify the names of the inputs specified in your model under
input_binding_names. In the case of ONNX models converted from PyTorch inputs names take the formINPUT__0.d. Specify the names of the inputs specified in your model under
output_binding_names. In the case of ONNX models converted from PyTorch and then thegraph_surgeon.pyconversion, names take the formoutput_old.Assuming the custom model input and output bindings are
MY_MODEL_INPUT_NAMEandMY_MODEL_OUTPUT_NAME, thenvidia::gxf::TensorRtInferencecomponent would result in:- type: nvidia::gxf::TensorRtInference parameters: input_binding_names: - MY_MODEL_INPUT_NAME output_binding_names: - MY_MODEL_OUTPUT_NAME
TipThe
nvidia::gxf::TensorRtInferencecomponent binds the names of the Holoscan component inputs to the model inputs via theinput_tensor_namesandinput_binding_nameslists, where the first specifies the name of the tensor used by the Holoscan componentnvidia::gxf::TensorRtInferenceand the latter specifies the name of the model input. Similarly,output_tensor_namesandoutput_binding_nameslink the component output names to the model output (see extensions).In the entity
segmentation_postprocessor, make the following change:network_output_type: sigmoid.In the entity
segmentation_visualizer, we will make the following changes undernvidia::holoscan::segmentation_visualizer::Visualizerto correctly input the dimensions of our video and the output dimensions of our model:a.
image_width: 720b.
image_height: 576c.
class_index_width: 512d.
class_index_height: 512Run the application with the new model and data.
cd /opt/nvidia/holoscan ./apps/ultrasound_segmentation/gxf/segmentation_replayer

Refer to the instructions in the Holoscan NGC container overview or the Github source repository to run the application, depending on how you’ve chosen to set up Holoscan.
The Multi AI application uses the models and data from iCardio.ai, and use the Multi AI Inference and Post processing operators among others to design the application. Those Multi AI operators use APIs from the Holoscan Inference module to extract data, initialize and execute the inference workflow, process and transmit data for visualization.

Multi AI sample application workflow with replay from file.
The pipeline uses a recorded ultrasound video file (generated by convert_video_to_gxf_entities script) for input frames.
Each input frame in the file is loaded by Video Stream Replayer and Broadcast node passes the frame to the following four nodes (Entities):
B-mode Perspective Preprocessor: Entity uses Format Converter to convert the data type of the image to
float32and resize the data to 320x240 per frame.Plax Chamber Resized: Entity uses Format Converter to resize the input image to 320x320x3 with
RGB888image format for visualization.Plax Chamber Preprocessor: Entity uses Format Converter to convert the data type of the image to
float32and resize the data to 320x320 per frame.Aortic Stenosis Preprocessor: Entity uses Format Converter to convert the data type of the image to
float32and resize the data to 300x300 per frame.
Then, Multi AI Inference uses outputs from three preprocessors to execute inference. Following inference, Multi AI Postprocessor uses inferred output to process as per specifications. Visualizer iCardio extension is then used to generate visualization components for the plax chamber output, which is then fed into HoloViz to generate the visualization. Output from the sample application demonstrates 5 keypoints identified by the Plax Chamber model. Keypoints are connected in the output frame as shown in the image below.

Aortic Stenosis and B-mode Perspective models are the Classification models. Classification results can be printed using print keyword against the output tensors from the Classification models in the Multi AI Postprocessor settings. Printing of the results is optional and can be ignored by removing relevant entries in the post processor settings.
Holoscan SDK provides capability to process all models in ONNX, TensorRT FP32 and TensorRT FP16 format. Classification models (Aortic Stenosis and B-mode Perspective), do not support TensorRT FP16 conversion. Plax Chamber model is supported for all available formats (ONNX, TensorRT FP32 and TensorRT FP16).
Bring Your Own Model (BYOM) - Customizing the Multi AI Application For Your Model
This section shows how the user can create a multi AI application using The Holoscan Inference component. The section will list down the steps to create the inference component of the pipeline, the user must write the pre-processing and post-processing steps and link them appropriately in the pipeline.
The user can use the
multiai_inferencedictionary in the application config atapps/multiai/cpp/app_config.yaml.The user has to populate all the elements in the dictionary as per the new set of models that user is bringing.
All models must be either in
onnxor intensorrt engine fileformat. User has to updatemodel_path_mapwith key as unique string and the value as the path to the model file on disk. The Holoscan Inference component will do theonnxtotensorrtmodel conversion if the converted files do not exist and if the user has set the parameters for the same.Keys in
model_path_map,pre_processor_mapandinference_mapmust be the same, representing each of the models.pre_processor_mapcontains the key as a unique string representing each model, and the value as a list of the input tensors going into the model. Only one value per model is supported. Two or more models can have same input tensor name as their input.inference_mapcontains the key as a unique string representing each model, and the value as a string representing the tensor name of the output from the inference.in_tensor_namescontains the list of the input tensor names.out_tensor_namescontains the list of the output tensor names.The user can update the
backendas desired. The options areonnxrtandtrt. If the input models are intensorrt engine fileformat, then the user must selecttrtasbackend. If the input models are inonnxformat, then the user can select eithertrtoronnxas the backend.
Other parameters and the limitations are described in Parameters and Related Features section of the Holoscan Inference Component.
Once the inference component is created or updated, the user can execute the Multi AI application as described above in the Running the application section.