Accelerated DICOM decoding/encoding#
This notebook demonstrates how to manipulate DICOM files using nvImageCodec, particularly those that use compressed transfer syntaxes. nvImageCodec can efficiently decode compressed DICOM images, providing significant performance benefits for medical imaging workflows.
Prerequisites#
This notebook requires:
pydicom >= 3.0.0
pylibjpeg >= 2.0.0
pylibjpeg-openjpeg >= 2.0.0
pip install pydicom>=3.0.0
pip install pylibjpeg>=2.0.0
pip install pylibjpeg-openjpeg>=2.0.0
[1]:
try:
import pydicom
from packaging.version import parse
v = pydicom.__version__
print(f"pydicom version: {v}")
if parse(v) < parse("3.0.0"):
print("⚠️ Please upgrade pydicom to >=3.0.0")
else:
print("✓ pydicom version is compatible")
except ImportError:
print("❌ pydicom is not installed.\nPlease install: pip install pydicom>=3.0.0")
pydicom version: 3.0.1
✓ pydicom version is compatible
Let’s run nvidia-smi to verify the GPU available on this system
[2]:
! nvidia-smi
Fri Nov 14 11:22:14 2025
+-----------------------------------------------------------------------------------------+
| NVIDIA-SMI 580.40 Driver Version: 580.40 CUDA Version: 13.0 |
+-----------------------------------------+------------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+========================+======================|
| 0 NVIDIA A100-PCIE-40GB On | 00000000:65:00.0 Off | 0 |
| N/A 30C P0 33W / 250W | 425MiB / 40960MiB | 0% Default |
| | | Disabled |
+-----------------------------------------+------------------------+----------------------+
+-----------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=========================================================================================|
+-----------------------------------------------------------------------------------------+
Load DICOM Series with pydicom#
We will be using DICOM data from the assets/DICOM/MRI/Knee directory, which is originally taken from the DALI_extra repository.
Let’s load the DICOM series SER00001 and examine its metadata.
[3]:
import os
import glob
import time
# Path to DICOM series
resources_dir = os.getenv("PYNVIMGCODEC_EXAMPLES_RESOURCES_DIR", "../assets/images")
dicom_series_dir = os.path.join(resources_dir, "DICOM/MRI/Knee/DICOMS/STU00001/SER00001")
# Get all DICOM files in the series
dicom_files = sorted(glob.glob(os.path.join(dicom_series_dir, "*.dcm")))
print(f"Found {len(dicom_files)} DICOM files in series {dicom_series_dir}")
print(f"\nFirst file: {os.path.basename(dicom_files[0])}")
print(f"Last file: {os.path.basename(dicom_files[-1])}")
print(f"Loading {len(dicom_files)} DICOM files...")
start_time = time.time()
dicom_series = [pydicom.dcmread(f) for f in dicom_files]
dcmread_time = time.time() - start_time
# Mention the original transfer syntax from the first DICOM file
original_transfer_syntax = dicom_series[0].file_meta.TransferSyntaxUID.name
print(f"✓ Loaded {len(dicom_series)} files in {dcmread_time:.4f}s ({dcmread_time/len(dicom_files)*1000:.2f} ms/file)")
print(f"Transfer syntax: {original_transfer_syntax}")
print(f"Volume shape: {dicom_series[0].Rows} x {dicom_series[0].Columns} x {len(dicom_series)}")
# Show metadata of the first DICOM file
first_dicom = dicom_series[0]
print("=" * 70)
print("DICOM Metadata")
print("=" * 70)
print(f"Modality: {getattr(first_dicom, 'Modality', 'N/A')}")
print(f"Study: {getattr(first_dicom, 'StudyDescription', 'N/A')}")
print(f"Series: {getattr(first_dicom, 'SeriesDescription', 'N/A')}")
print(f"\nImage Dimensions: {first_dicom.Rows} x {first_dicom.Columns}")
print(f"Bits Stored: {first_dicom.BitsStored} / {first_dicom.BitsAllocated}")
print(f"\nTransfer Syntax: {first_dicom.file_meta.TransferSyntaxUID.name}")
print(f"Is Compressed: {first_dicom.file_meta.TransferSyntaxUID.is_compressed}")
Found 38 DICOM files in series ../../../example/assets/images/DICOM/MRI/Knee/DICOMS/STU00001/SER00001
First file: IMG00001.dcm
Last file: IMG00038.dcm
Loading 38 DICOM files...
✓ Loaded 38 files in 0.0364s (0.96 ms/file)
Transfer syntax: Implicit VR Little Endian
Volume shape: 234 x 256 x 38
======================================================================
DICOM Metadata
======================================================================
Modality: MR
Study: JOINTS^KNEE RIGHT
Series: t1_tse_tra
Image Dimensions: 234 x 256
Bits Stored: 12 / 16
Transfer Syntax: Implicit VR Little Endian
Is Compressed: False
Now that we’ve loaded the DICOM files and their metadata, note that the pixel data itself has not yet been loaded into memory. With pydicom, accessing the .pixel_array property will read and decode the pixel data on demand, automatically selecting the correct decoding plugin depending on the file’s transfer syntax.
Before decoding the pixel data, we need to ensure the slices are in the correct spatial order. DICOM files may not always be stored in sequential order on disk, so we’ll sort them based on their position using metadata fields such as InstanceNumber, ImagePositionPatient, or SliceLocation. This ensures the reconstructed 3D volume has slices in the proper anatomical order.
[4]:
import numpy as np
# Sort slices by Instance Number or Image Position Patient
print("Sorting slices by slice position...")
# Try to sort by Instance Number first (most common)
if hasattr(dicom_series[0], 'InstanceNumber'):
dicom_series_sorted = sorted(dicom_series, key=lambda x: x.InstanceNumber)
print("Sorted by Instance Number")
# If Instance Number is not available, sort by Image Position Patient (slice location)
elif hasattr(dicom_series[0], 'ImagePositionPatient'):
dicom_series_sorted = sorted(dicom_series, key=lambda x: float(x.ImagePositionPatient[2]))
print("Sorted by Image Position Patient (Z coordinate)")
# Fallback: sort by Slice Location if available
elif hasattr(dicom_series[0], 'SliceLocation'):
dicom_series_sorted = sorted(dicom_series, key=lambda x: float(x.SliceLocation))
print("Sorted by Slice Location")
else:
# Keep original order if no sorting criteria available
dicom_series_sorted = dicom_series
print("Warning: No suitable sorting criteria found, keeping original order")
print(f"\nFirst slice Instance Number: {getattr(dicom_series_sorted[0], 'InstanceNumber', 'N/A')}")
print(f"Last slice Instance Number: {getattr(dicom_series_sorted[-1], 'InstanceNumber', 'N/A')}")
# Decode pixel data to numpy array
print("\nDecoding pixel data to numpy array...")
# Time the pixel_array access loop
start_time = time.time()
pixel_arrays = []
for ds in dicom_series_sorted:
pixel_arrays.append(ds.pixel_array)
volume = np.stack(pixel_arrays)
pixel_decode_time = time.time() - start_time
print(f"⏱️ Pixel data decode time: {pixel_decode_time:.4f} seconds ({pixel_decode_time/len(dicom_series_sorted)*1000:.2f} ms per slice)")
print(f"\nVolume shape: {volume.shape}")
print(f"Volume dtype: {volume.dtype}")
print(f"Volume range: [{volume.min()}, {volume.max()}]")
print(f"Memory size: {volume.nbytes / (1024**2):.2f} MB")
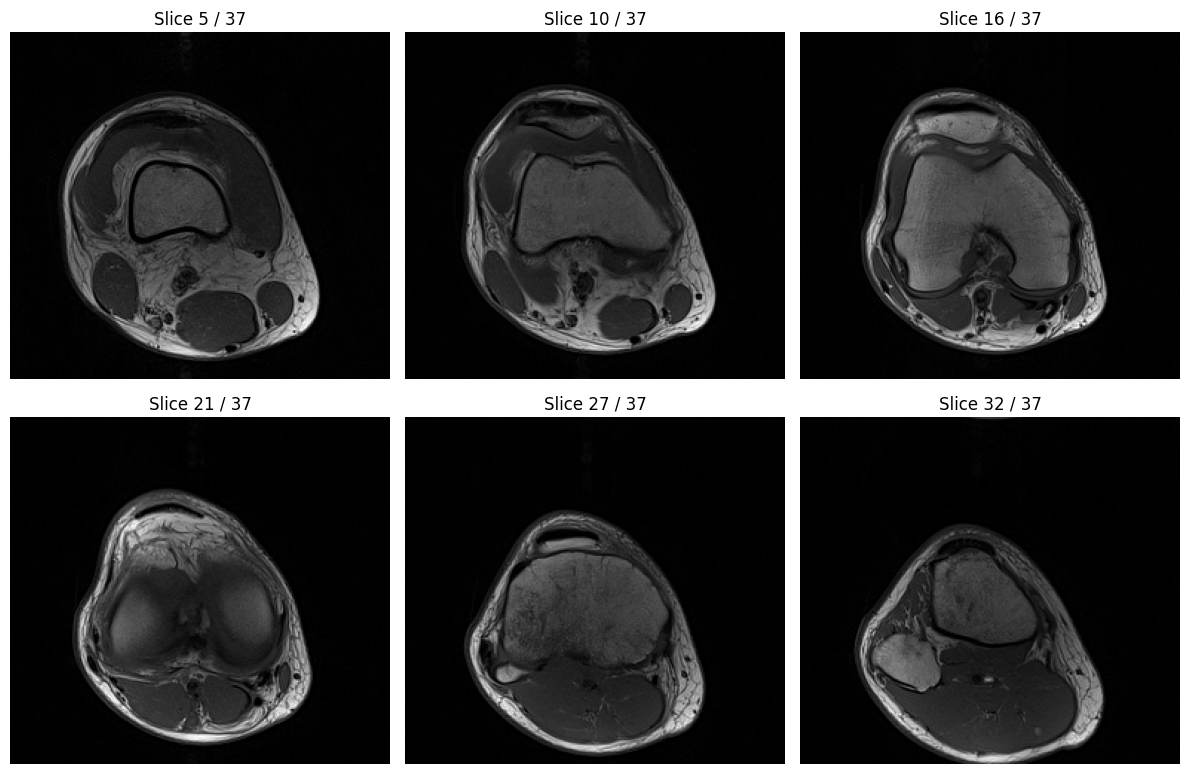
import matplotlib.pyplot as plt
# Visualize a few representative slices from the volume
fig, axes = plt.subplots(2, 3, figsize=(12, 8))
axes = axes.flatten()
# Select 6 evenly spaced slices to display
num_slices = volume.shape[0]
slice_indices = [int(i * num_slices / 7) for i in range(1, 7)]
for idx, ax in enumerate(axes):
slice_idx = slice_indices[idx]
ax.imshow(volume[slice_idx], cmap="gray")
ax.set_title(f"Slice {slice_idx} / {num_slices-1}")
ax.axis("off")
plt.tight_layout()
plt.show()
Sorting slices by slice position...
Sorted by Instance Number
First slice Instance Number: 1
Last slice Instance Number: 38
Decoding pixel data to numpy array...
⏱️ Pixel data decode time: 0.0288 seconds (0.76 ms per slice)
Volume shape: (38, 234, 256)
Volume dtype: uint16
Volume range: [0, 2139]
Memory size: 4.34 MB
Transcode DICOM to HTJ2K with nvImageCodec#
The original DICOM files use uncompressed pixel data (Implicit VR Little Endian), which is simple but inefficient for storage and transfer. Let’s transcode them to HTJ2K (High-Throughput JPEG 2000), a modern compression standard that offers:
Lossless compression: No quality loss, critical for medical imaging
Better compression ratios: ~2-3x smaller files than uncompressed
Hardware-accelerated decoding: Faster decompression on GPU
Standardized: Official DICOM transfer syntax (UID 1.2.840.10008.1.2.4.201)
HTJ2K is particularly valuable for medical imaging archives, PACS systems, and telemedicine applications where storage costs and network bandwidth are concerns.
[5]:
from nvidia import nvimgcodec
import tempfile
import shutil
from pydicom.uid import JPEG2000Lossless
# Create encoder instance
encoder = nvimgcodec.Encoder()
# Create temporary directory for HTJ2K transcoded files
htj2k_dir = tempfile.mkdtemp(prefix="dicom_htj2k_")
print(f"Created temporary directory: {htj2k_dir}")
# HTJ2K Transfer Syntax UID
HTJ2K_LOSSLESS_UID = "1.2.840.10008.1.2.4.201" # High-Throughput JPEG 2000 Image Compression (Lossless Only)
print(f"\nOriginal Transfer Syntax: {dicom_series_sorted[0].file_meta.TransferSyntaxUID.name}")
print(f"Target Transfer Syntax: HTJ2K Lossless")
Created temporary directory: /tmp/dicom_htj2k_ncr__yd6
Original Transfer Syntax: Implicit VR Little Endian
Target Transfer Syntax: HTJ2K Lossless
[6]:
# Transcode each DICOM file to HTJ2K
print("\n" + "=" * 70)
print(f"Transcoding {len(dicom_series_sorted)} DICOM files to HTJ2K...")
print("=" * 70)
# Setup encode parameters for HTJ2K lossless
jpeg2k_encode_params = nvimgcodec.Jpeg2kEncodeParams()
jpeg2k_encode_params.num_resolutions = 6 # good balance
jpeg2k_encode_params.code_block_size = (64, 64)
jpeg2k_encode_params.bitstream_type = nvimgcodec.Jpeg2kBitstreamType.J2K
jpeg2k_encode_params.prog_order = nvimgcodec.Jpeg2kProgOrder.LRCP
jpeg2k_encode_params.ht = True # Enable High Throughput mode
encode_params = nvimgcodec.EncodeParams(
quality_type=nvimgcodec.QualityType.LOSSLESS,
jpeg2k_encode_params=jpeg2k_encode_params
)
start_time = time.time()
encoded = [bytes(x) for x in encoder.encode(pixel_arrays, "jpeg2k", params=encode_params)]
total_encode_time = time.time() - start_time
from pydicom.dataset import Dataset, FileMetaDataset
from pydicom.uid import ExplicitVRLittleEndian
from pydicom.encaps import encapsulate
compressed_total_size = 0
original_total_size = 0
htj2k_dicom_files = []
print("Saving HTJ2K-encoded DICOM files...")
for i, ds in enumerate(dicom_series_sorted):
# Clone the original dataset to preserve all metadata
new_ds = ds.copy()
# Update Transfer Syntax UID in file_meta
new_ds.file_meta.TransferSyntaxUID = HTJ2K_LOSSLESS_UID
new_ds.file_meta.is_implicit_VR = False
new_ds.file_meta.is_little_endian = True
# Encapsulate the HTJ2K encoded data properly for DICOM
# encapsulate() expects a list of frame fragments
new_ds.PixelData = encapsulate([encoded[i]])
# (Optional) update Instance Creation Date/Time for reproducibility
from datetime import datetime
new_ds.InstanceCreationDate = datetime.now().strftime('%Y%m%d')
new_ds.InstanceCreationTime = datetime.now().strftime('%H%M%S.%f')[:18]
# Save to new directory with the same filename as the original
original_filename = os.path.basename(ds.filename)
out_path = os.path.join(htj2k_dir, original_filename)
new_ds.save_as(out_path)
htj2k_dicom_files.append(out_path)
# Check the size of the files on disk (after saving)
compressed_total_size += os.path.getsize(out_path)
original_total_size += os.path.getsize(ds.filename)
print("\n========== HTJ2K Encoding Summary ==========")
print(f"• Number of slices encoded : {len(dicom_series_sorted)}")
print(f"• Total encoding time : {total_encode_time:.4f} seconds ({total_encode_time/len(dicom_series_sorted)*1000:.2f} ms/slice)")
print("\n📊 Compression Results:")
print(f" - Original total size : {original_total_size / 1024:.2f} KB ({original_total_size / (1024**2):.2f} MB)")
print(f" - Compressed total size : {compressed_total_size / 1024:.2f} KB ({compressed_total_size / (1024**2):.2f} MB)")
if compressed_total_size > 0:
print(f" - Compression ratio : {original_total_size / compressed_total_size:.2f}x smaller")
print(f" - Space saved : {(original_total_size - compressed_total_size) / 1024:.2f} KB ({(1 - compressed_total_size/original_total_size)*100:.1f}%)")
else:
print(" - Compression ratio : N/A (compressed size is zero)")
print(" - Space saved : N/A")
print(f"\n✓ All HTJ2K-encoded DICOM files are saved at: {htj2k_dir}")
======================================================================
Transcoding 38 DICOM files to HTJ2K...
======================================================================
Saving HTJ2K-encoded DICOM files...
========== HTJ2K Encoding Summary ==========
• Number of slices encoded : 38
• Total encoding time : 0.0508 seconds (1.34 ms/slice)
📊 Compression Results:
- Original total size : 4506.39 KB (4.40 MB)
- Compressed total size : 1883.03 KB (1.84 MB)
- Compression ratio : 2.39x smaller
- Space saved : 2623.37 KB (58.2%)
✓ All HTJ2K-encoded DICOM files are saved at: /tmp/dicom_htj2k_ncr__yd6
Verify HTJ2K Decoding Performance#
Now let’s verify the transcoded HTJ2K files can be decoded correctly, and compare decoding performance between pydicom (CPU-based) and nvImageCodec (GPU-accelerated).
[7]:
# Parse HTJ2K files
datasets = [pydicom.dcmread(f) for f in htj2k_dicom_files]
# Method 1: pydicom pixel_array (CPU-based decoding)
start_time = time.time()
pydicom_decoded = [ds.pixel_array for ds in datasets]
pydicom_time = time.time() - start_time
# Method 2: nvImageCodec (GPU-accelerated decoding)
decoder = nvimgcodec.Decoder()
decode_params = nvimgcodec.DecodeParams(allow_any_depth=True, color_spec=nvimgcodec.ColorSpec.UNCHANGED)
# Extract encoded frame data from DICOM PixelData
encoded_frames = []
for ds in datasets:
encoded_frames.extend(pydicom.encaps.generate_frames(ds.PixelData))
start_time = time.time()
nvimg_decoded = decoder.decode(encoded_frames, params=decode_params)
nvimg_time = time.time() - start_time
nvimg_decoded_cpu = [np.array(img.cpu()) for img in nvimg_decoded]
print(f"⏱️ pydicom decode time: {pydicom_time:.4f}s ({pydicom_time/len(datasets)*1000:.2f} ms/file)")
print(f"⏱️ nvImageCodec decode time: {nvimg_time:.4f}s ({nvimg_time/len(datasets)*1000:.2f} ms/file)")
print(f"🚀 Speedup: {pydicom_time/nvimg_time:.2f}x faster")
# Verify correctness
for idx in range(len(nvimg_decoded_cpu)):
d = nvimg_decoded_cpu[idx].squeeze()
assert np.array_equal(d, volume[idx]), f"Slice {idx} mismatch"
print("✓ All decoded slices match original")
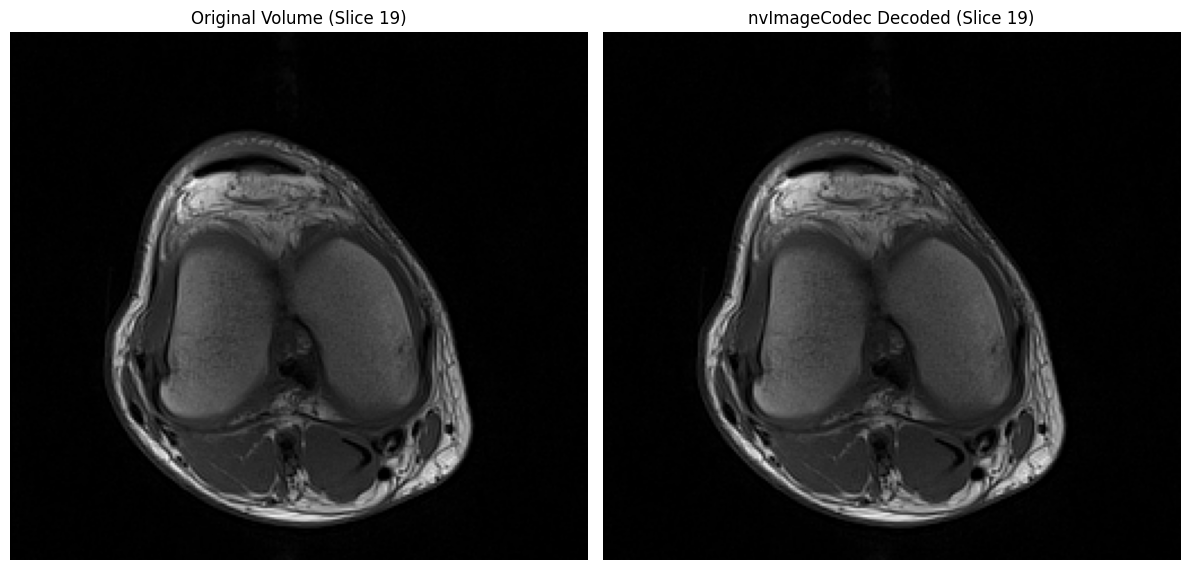
# Visual comparison
mid_idx = len(volume) // 2
fig, axs = plt.subplots(1, 2, figsize=(12, 6))
axs[0].imshow(volume[mid_idx], cmap='gray')
axs[0].set_title('Original Volume (Slice {})'.format(mid_idx))
axs[0].axis('off')
axs[1].imshow(nvimg_decoded_cpu[mid_idx], cmap='gray')
axs[1].set_title('nvImageCodec Decoded (Slice {})'.format(mid_idx))
axs[1].axis('off')
plt.tight_layout()
plt.show()
⏱️ pydicom decode time: 0.0675s (1.78 ms/file)
⏱️ nvImageCodec decode time: 0.0367s (0.96 ms/file)
🚀 Speedup: 1.84x faster
✓ All decoded slices match original
Multi-Frame DICOM#
So far we’ve been working with single-frame DICOM files (one file per slice). However, DICOM also supports multi-frame images, where multiple slices are stored in a single file. This approach offers several advantages:
Reduced filesystem overhead: One file instead of 38 means fewer inodes, faster directory operations
Slightly better compression: Shared metadata reduces redundancy
Simplified file management: Easier to archive, transfer, and track
Let’s convert our 38 single-frame HTJ2K files into a single multi-frame HTJ2K DICOM file.
[8]:
import pydicom.uid
from pydicom.encaps import encapsulate
# Create multi-frame DICOM from single-frame HTJ2K files
output_ds = datasets[0].copy()
output_ds.SOPInstanceUID = output_ds.SeriesInstanceUID
# Collect all encoded frames
all_encoded_frames = []
for ds in datasets:
frames = list(pydicom.encaps.generate_frames(ds.PixelData))
all_encoded_frames.extend(frames)
# Encapsulate as multi-frame
output_ds.PixelData = encapsulate(all_encoded_frames, has_bot=True)
output_ds.NumberOfFrames = len(all_encoded_frames)
# Save
multiframe_htj2k_dir = tempfile.mkdtemp(prefix="multiframe_htj2k_")
multi_frame_file = os.path.join(multiframe_htj2k_dir, "multiframe_htj2k.dcm")
output_ds.save_as(multi_frame_file)
print(f"✓ Created multi-frame DICOM: {multi_frame_file}")
# Size comparison
original_size = sum(os.path.getsize(f) for f in dicom_files)
single_htj2k_size = sum(os.path.getsize(f) for f in htj2k_dicom_files)
multi_htj2k_size = os.path.getsize(multi_frame_file)
print(f"\n📦 File Size Comparison:")
print(f" Original (38 files): {original_size/1024**2:.2f} MB")
print(f" Single-frame HTJ2K (38): {single_htj2k_size/1024**2:.2f} MB ({original_size/single_htj2k_size:.2f}x smaller)")
print(f" Multi-frame HTJ2K (1): {multi_htj2k_size/1024**2:.2f} MB ({original_size/multi_htj2k_size:.2f}x smaller)")
✓ Created multi-frame DICOM: /tmp/multiframe_htj2k_62_f9cgt/multiframe_htj2k.dcm
📦 File Size Comparison:
Original (38 files): 4.40 MB
Single-frame HTJ2K (38): 1.84 MB (2.39x smaller)
Multi-frame HTJ2K (1): 1.78 MB (2.47x smaller)
[9]:
# Verify multi-frame DICOM with pydicom
mf_ds = pydicom.dcmread(multi_frame_file)
start_time = time.time()
mf_arr = mf_ds.pixel_array
pydicom_time = time.time() - start_time
print(f"⏱️ pydicom multi-frame decode time: {pydicom_time:.4f}s ({pydicom_time/len(mf_arr)*1000:.2f} ms/frame)")
print(f"Multi-frame shape: {mf_arr.shape}")
print(f"Transfer Syntax: {mf_ds.file_meta.TransferSyntaxUID.name}")
# Verify all frames match original
orig_stack = np.stack([ds.pixel_array for ds in dicom_series_sorted], axis=0)
assert np.array_equal(orig_stack, mf_arr), "Multi-frame data mismatch!"
print("✓ All frames match original")
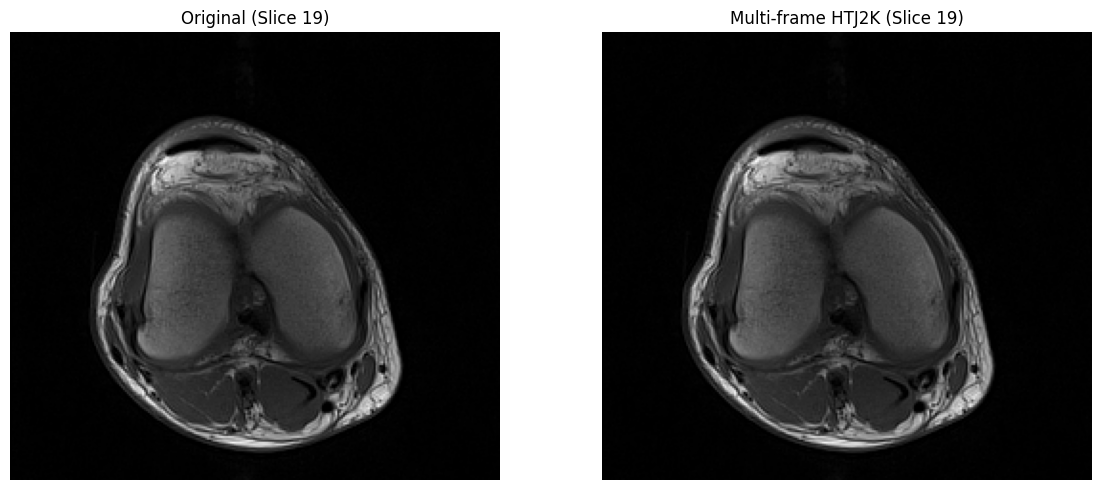
# Visual comparison
mid_idx = len(mf_arr) // 2
fig, axs = plt.subplots(1, 2, figsize=(12, 5))
axs[0].imshow(orig_stack[mid_idx], cmap='gray')
axs[0].set_title(f"Original (Slice {mid_idx})")
axs[0].axis('off')
axs[1].imshow(mf_arr[mid_idx], cmap='gray')
axs[1].set_title(f"Multi-frame HTJ2K (Slice {mid_idx})")
axs[1].axis('off')
plt.tight_layout()
plt.show()
⏱️ pydicom multi-frame decode time: 0.0577s (1.52 ms/frame)
Multi-frame shape: (38, 234, 256)
Transfer Syntax: High-Throughput JPEG 2000 Image Compression (Lossless Only)
✓ All frames match original
[10]:
# Decode multi-frame DICOM with nvImageCodec
print("\nDecoding multi-frame HTJ2K with nvImageCodec...")
encoded_frames = list(pydicom.encaps.generate_frames(mf_ds.PixelData))
start_time = time.time()
decoded_frames = decoder.decode(encoded_frames, params=decode_params)
nvimg_time = time.time() - start_time
decoded_arrays = [np.array(x.cpu()).squeeze() for x in decoded_frames]
decoded_stack = np.stack(decoded_arrays, axis=0)
print(f"⏱️ Decode time: {nvimg_time:.4f}s ({nvimg_time/len(decoded_frames)*1000:.2f} ms/frame)")
speedup = pydicom_time / nvimg_time
print(f"🚀 Speedup vs pydicom: {speedup:.2f}x faster with nvImageCodec")
# Verify correctness
assert np.array_equal(decoded_stack, orig_stack), "nvImageCodec decode mismatch!"
print("✓ nvImageCodec multi-frame decode matches original")
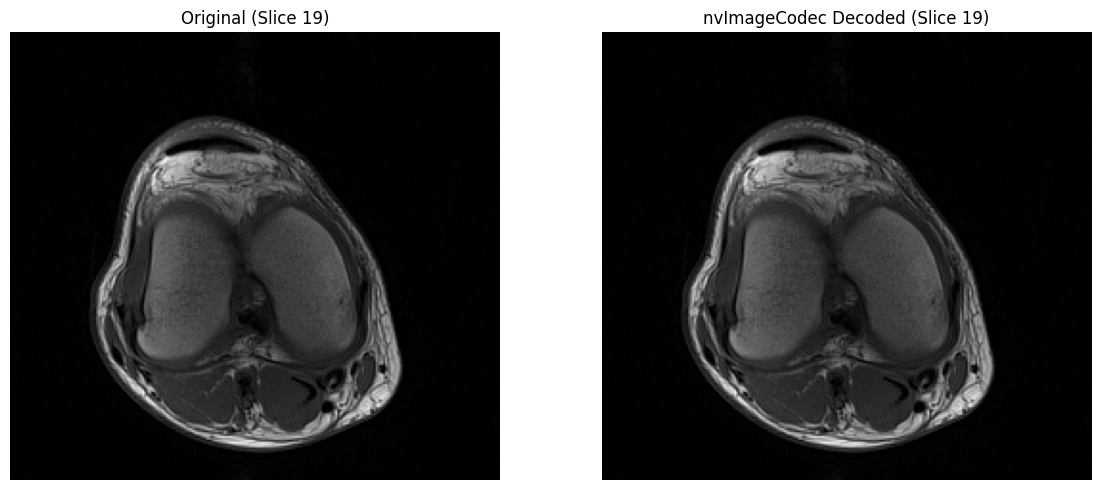
# Visual comparison
mid_idx = len(decoded_stack) // 2
fig, axs = plt.subplots(1, 2, figsize=(12, 5))
axs[0].imshow(orig_stack[mid_idx], cmap='gray')
axs[0].set_title(f"Original (Slice {mid_idx})")
axs[0].axis('off')
axs[1].imshow(decoded_stack[mid_idx], cmap='gray')
axs[1].set_title(f"nvImageCodec Decoded (Slice {mid_idx})")
axs[1].axis('off')
plt.tight_layout()
plt.show()
Decoding multi-frame HTJ2K with nvImageCodec...
⏱️ Decode time: 0.0208s (0.55 ms/frame)
🚀 Speedup vs pydicom: 2.78x faster with nvImageCodec
✓ nvImageCodec multi-frame decode matches original
[11]:
# Cleanup temporary directories
import shutil
shutil.rmtree(htj2k_dir, ignore_errors=True)
shutil.rmtree(multiframe_htj2k_dir, ignore_errors=True)
print("✓ Cleaned up temporary directories")
✓ Cleaned up temporary directories
[12]:
print("\n" + "=" * 80)
print("SUMMARY")
print("=" * 80)
print(f"\n📊 Compression Results:")
print(f" • Original size: {original_size/1024**2:.2f} MB (38 uncompressed files)")
print(f" • HTJ2K single-frame: {single_htj2k_size/1024**2:.2f} MB ({original_size/single_htj2k_size:.2f}x smaller, 38 files)")
print(f" • HTJ2K multi-frame: {multi_htj2k_size/1024**2:.2f} MB ({original_size/multi_htj2k_size:.2f}x smaller, 1 file)")
print(f"\n🎯 Key Takeaways:")
print(f" • HTJ2K provided ~{original_size/single_htj2k_size:.1f}x lossless compression for our dataset")
print(f" • nvImageCodec accelerated HTJ2K decoding on GPU, providing ~{speedup:.2f}x speedup over default pydicom decoding")
print(f" • Multi-frame format reduces file count and overhead")
print(f" • All methods preserve image quality (lossless)")
================================================================================
SUMMARY
================================================================================
📊 Compression Results:
• Original size: 4.40 MB (38 uncompressed files)
• HTJ2K single-frame: 1.84 MB (2.39x smaller, 38 files)
• HTJ2K multi-frame: 1.78 MB (2.47x smaller, 1 file)
🎯 Key Takeaways:
• HTJ2K provided ~2.4x lossless compression for our dataset
• nvImageCodec accelerated HTJ2K decoding on GPU, providing ~2.78x speedup over default pydicom decoding
• Multi-frame format reduces file count and overhead
• All methods preserve image quality (lossless)