Important
You are viewing the NeMo 2.0 documentation. This release introduces significant changes to the API and a new library, NeMo Run. We are currently porting all features from NeMo 1.0 to 2.0. For documentation on previous versions or features not yet available in 2.0, please refer to the NeMo 24.07 documentation.
Entity Linking#
Entity linking is the process of matching concepts mentioned in natural language to their unique IDs and canonical forms stored
in a knowledge base. For example, an entity linking model might match the phrase blood thinners mentioned in conversation
to the knowledge base concept UID45623 anticoagulant. Entity linking applications range from helping automate ingestion of
large amounts of data to assisting in real time concept normalization.
Within NeMo we use the entity linking approach described in Liu et. al’s NAACL 2021 “Self-alignment Pre-training for Biomedical Entity Representations” [nlp-entity_linking1]. The main idea behind this approach is to reshape an initial concept embedding space such that synonyms of the same concept are pulled closer together and unrelated concepts are pushed further apart. The concept embeddings from this reshaped space can then be used to build a knowledge base embedding index.
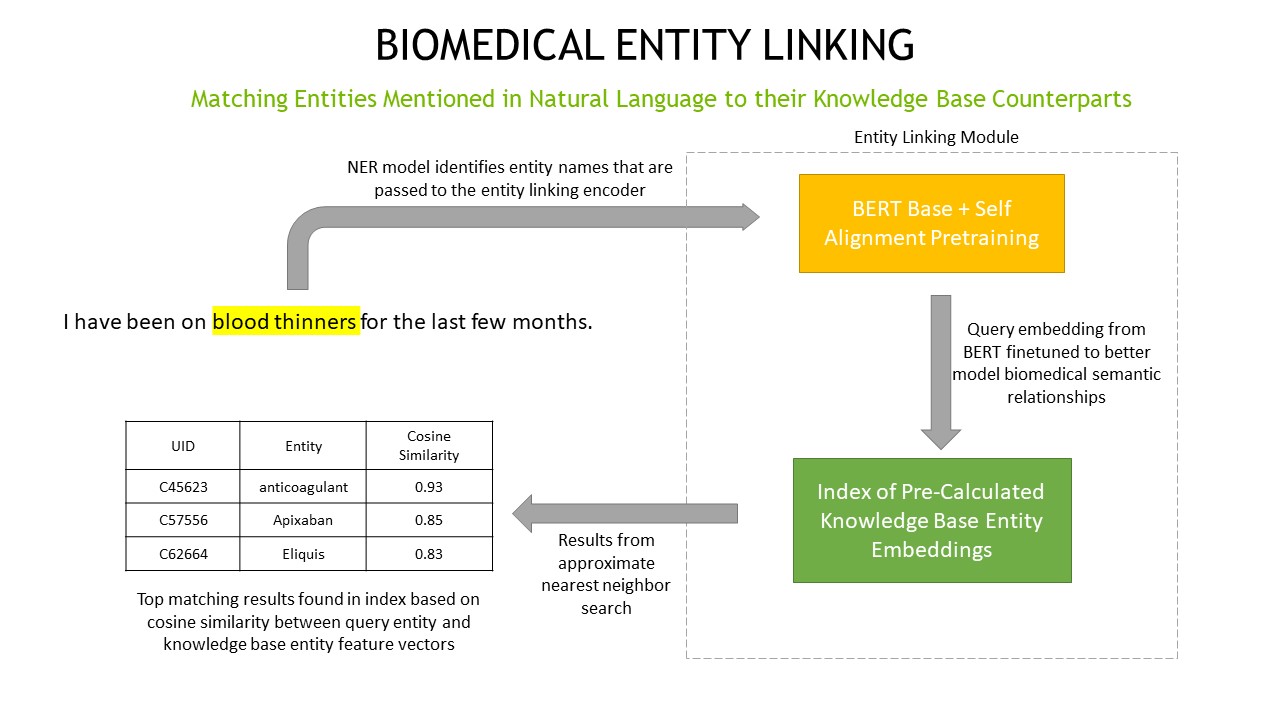
Our BERT-base + Self Alignment Pretraining implementation allows you to train an entity linking encoder. We also provide example code on building an index with Medical UMLS concepts NeMo/examples/nlp/entity_linking/build_index.py.
Please try the example Entity Linking model in a Jupyter notebook (can run on Google’s Colab).
Connect to an instance with a GPU (Runtime -> Change runtime type -> select GPU for the hardware accelerator).
An example script on how to train the model can be found here: NeMo/examples/nlp/entity_linking.
References#
Fangyu Liu, Ehsan Shareghi, Zaiqiao Meng, Marco Basaldella, and Nigel Collier. Self-alignment pretraining for biomedical entity representations. 2021. arXiv:2010.11784.