EquiDock
Contents
EquiDock#
Model Overview#
Description:#
EquiDock [1] predicts protein-protein complex formation from two individual proteins.
EquiDock is an SE(3)-equivariant model that treats both the proteins as rigid bodies and assumes no conformational change during the formation of the complex. This model is ready for commercial use.
Third-Party Community Consideration#
The original model was not owned or developed by NVIDIA. This model has been developed and built to a third-party’s requirements for this application and use case; see GitHub for more.
References:#
Ganea, Octavian-Eugen, Xinyuan Huang, Charlotte Bunne, Yatao Bian, Regina Barzilay, Tommi Jaakkola, and Andreas Krause. “Independent SE(3)-equivariant models for end-to-end rigid protein docking.” arXiv preprint arXiv:2111.07786 (2021).
Kabsch, Wolfgang. “A solution for the best rotation to relate two sets of vectors.” Acta Crystallographica Section A: Crystal Physics, Diffraction, Theoretical and General Crystallography 32, no. 5 (1976): 922-923.
Vreven, Thom, Iain H. Moal, Anna Vangone, Brian G. Pierce, Panagiotis L. Kastritis, Mieczyslaw Torchala, Raphael Chaleil et al. “Updates to the integrated protein–protein interaction benchmarks: docking benchmark version 5 and affinity benchmark version 2.” Journal of molecular biology 427, no. 19 (2015): 3031-3041.
Townshend, Raphael, Rishi Bedi, Patricia Suriana, and Ron Dror. “End-to-end learning on 3d protein structure for interface prediction.” Advances in Neural Information Processing Systems 32 (2019).
Model Architecture:#
Architecture Type: Graph Neural Network (GNN)
Network Architecture: Independent SE(3)-Equivariant Graph Matching Network (IEGMN)
Input:#
Input Type(s): Text (Geometric Protein Structure)
Input Format(s): Binary File
Other Properties Related to Input: Text represented as integers and floating point 32, and alphabetical representations for the residue name. Maximum number of residues is 400 per protein and maximum number of atoms is 4000 per protein
Output:#
Output Types: Text (Geometric Protein Structure)
Output Format(s): Protein Data Bank (PDB)
Other Properties Related to Output: [ Text represented as integers, floating point 32, and alphabetical representations for the residue name.]
Software Integration:#
Runtime Engine(s):
BioNeMo, NeMo 1.2
Supported Hardware Microarchitecture Compatibility:
[Ampere]
[Hopper]
[Preferred/Supported] Operating System(s):
[Linux]
Model Version(s):#
dips.23.10
db5.23.10
Training & Evaluation:#
Training Dataset:#
Link: https://zlab.umassmed.edu/benchmark/ and https://github.com/drorlab/DIPS
Data Collection Method by dataset
[Automatic/Sensors]
Labeling Method by dataset[Not Applicable]
Properties:
DB5.5 dataset consists of 253 protein structures built by mining the Protein Data Bank (PDB) for pairs of interacting proteins The Database for Interacting Proteins Structures (DIPS) has 41,876 binary complexes containing bound structures with rigid body docking, while DB5.5 includes unbound protein structures. Datasets are then randomly partitioned in training, validation, and testing datasets The training and validation are used during training. DB5.5 includes 203/25 training and validation data points respectively. DIPS includes 39,937/974 training and validation data points respectively.
Dataset License(s): [https://jirasw.nvidia.com/browse/DGPTT-1300, https://jirasw.nvidia.com/browse/DGPTT-1301] INTERNAL ONLY
Evaluation Dataset:#
Link: https://zlab.umassmed.edu/benchmark/ and https://github.com/drorlab/DIPS
Data Collection Method by dataset
[Automatic/Sensors]
Labeling Method by dataset[Not Applicable]
Properties:
DB5.5 dataset consists of 253 protein structures built by mining the Protein Data Bank (PDB) for pairs of interacting proteins The Database for Interacting Proteins Structures (DIPS) has 41,876 binary complexes containing bound structures with rigid body docking, while DB5.5 includes unbound protein structures.
Datasets are then randomly partitioned in training, validation, and testing datasets The training and validation are used during training. DB5.5 and DIPS testing datasets include 25 and 965 testing data points respectively.
Dataset License(s): [https://jirasw.nvidia.com/browse/DGPTT-1300, https://jirasw.nvidia.com/browse/DGPTT-1301] INTERNAL ONLY
Ethical Considerations:#
NVIDIA believes Trustworthy AI is a shared responsibility and we have established policies and practices to enable development for a wide array of AI applications. When downloaded or used in accordance with our terms of service, developers should work with their team to ensure this model meets requirements for the relevant industry and use case and addresses unforeseen product misuse. For more detailed information on ethical considerations for this model, please see the Model Card++ Explainability, Bias, Safety & Security, and Privacy Subcards [Insert Link to Model Card++ here]. Please report security vulnerabilities or NVIDIA AI Concerns here.
Benchmarks#
Two pretrained EquiDock checkpoints are available for download: equidock_dips.nemo and equidock_db5.nemo. These have been converted to the NeMo format from publicly available checkpoints. This section provides accuracy benchmarks for these models, as well as information on expected training speed performance. Currently, models trained from randomly initialized weights within the BioNeMo framework are not provided. The production of these models is ongoing work.
Accuracy Benchmarks#
The accuracy of EquiDock was measured using the complex and interface root-mean-squared distance [Ganea et al., 2021], which are measured over test datasets of DIPS and DB5.
RMSD type |
Checkpoint Name |
Dataset |
Dataset Split for Measurement |
Metric |
BioNeMo |
Public |
|---|---|---|---|---|---|---|
Complex RMSD |
equidock_dips.nemo |
DIPS |
test |
median |
13.39 |
13.39 |
Complex RMSD |
equidock_dips.nemo |
DIPS |
test |
mean |
14.52 |
14.52 |
Complex RMSD |
equidock_dips.nemo |
DIPS |
test |
std |
7.13 |
7.13 |
Interface RMSD |
equidock_dips.nemo |
DIPS |
test |
median |
10.24 |
10.24 |
Interface RMSD |
equidock_dips.nemo |
DIPS |
test |
mean |
11.92 |
11.92 |
Interface RMSD |
equidock_dips.nemo |
DIPS |
test |
std |
7.01 |
7.01 |
Complex RMSD |
equidock_db5.nemo |
DB5 |
test |
median |
14.13 |
14.13 |
Complex RMSD |
equidock_db5.nemo |
DB5 |
test |
mean |
14.73 |
14.72 |
Complex RMSD |
equidock_db5.nemo |
DB5 |
test |
std |
5.31 |
5.31 |
Interface RMSD |
equidock_db5.nemo |
DB5 |
test |
median |
11.97 |
11.97 |
Interface RMSD |
equidock_db5.nemo |
DB5 |
test |
mean |
13.23 |
13.23 |
Interface RMSD |
equidock_db5.nemo |
DB5 |
test |
std |
4.93 |
4.93 |
Training Performance Benchmarks#
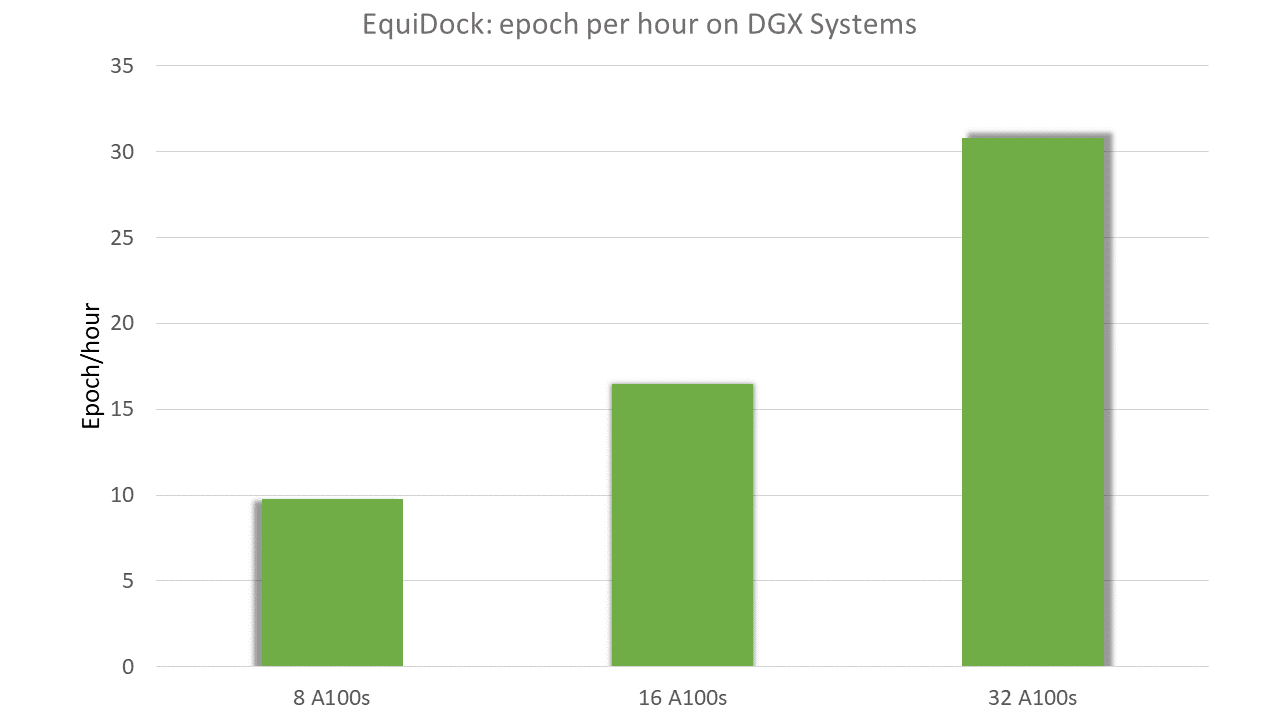
Training speed was tested on DGX-A100 systems with 8 IEGM layers over DIPS dataset for one epoch, on GPUs with 80GB of memory.
License#
EquiDock is provided under the Apache License.
