ESM-2nv
Contents
ESM-2nv#
Model Overview#
Description:#
ESM-2nv is a protein language model that provides numerical embeddings for each amino acid in a protein sequence. It is developed using the BioNeMo Framework. The embeddings from its encoder can be used as features for predictive models. The ESM-2nv 3B model has 36 layers, 40 attention heads, a hidden space dimension of 2560, and contains 3B parameters. The 650M model has 33 layers, 20 attention heads, a hidden space dimension of 1280, and contains 650M parameters. These models are ready for commercial use.
Third-Party Community Consideration#
This model is not owned or developed by NVIDIA. This model has been developed and built to a third-party’s requirements for this application and use case [1]; see link to Non-NVIDIA Model Card for ESM-2 3B model and non-NVIDIA Model Card for ESM-2 650M model
References:#
[1] Lin, Z., Akin, H., Rao, R., Hie, B., Zhu, Z., Lu, W., Smetanin, N., Verkuil, R., Kabeli, O., Shmueli, Y. and dos Santos Costa, A., 2023. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science, 379(6637), pp.1123-1130.
[2] “UniProt: the universal protein knowledgebase in 2021.” Nucleic acids research 49, no. D1 (2021): D480-D489.
[3] Devlin, J., Chang, M.W., Lee, K. and Toutanova, K., 2018. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
Model Architecture:#
Architecture Type: BERT
Network Architecture: ESM-2
Input:#
Input Type(s): Text (Protein Sequences)
Input Parameters: 1D
Other Properties Related to Input: Protein sequence represented as a string of canonical amino acids, of maximum length 1022. Longer sequences are automatically truncated to this length.
Output:#
Output Type(s): Text (Protein Sequences)
Output Parameters: 1D
Other Properties Related to Output: Numeric vector with one float-point value corresponding to an embedding for each amino acid in the input protein sequence. Maximum output length is 1022 embeddings - one embedding vector per amino acid.
Software Integration:#
Runtime Engine(s):
BioNeMo, NeMo 1.2
Supported Hardware Microarchitecture Compatibility:
[Ampere]
[Hopper]
[Volta]
[Preferred/Supported] Operating System(s):
[Linux]
Model Version(s):#
esm2nv_3B_converted.nemo, esm2nv_650M_converted.nemo, version 1.0
Training & Evaluation:#
Training Dataset:#
Data Collection Method by dataset
[Human]
Properties: UniRef50 (release 04/2021) was used for training [2]. The representative sequence for each UniRef50 cluster was selected, resulting in 49,874,565 protein sequences. The sequences were randomly split with 249,372 sequences in validation and 49,625,193 in training. All training sequences that matched a validation sequence with 50% sequence identity were removed from the train set, resulting in 49,425,807 train sequences. A sampling dataset of UniRef90 sequences was created based on any UniRef90 representatives and cluster members that had complete sequences available from UniRef90 or UniRef100, and filtered to UniRef90 sequences for clusters that corresponded to the UniRef50 train set. The UniRef90 dataset was combined with the filtered UniRef50 training dataset to create the sampling fasta file. A mapping file was created to enable rapid replacement of UniRef50 sequences with a sequence sampled uniformly from the corresponding records in the sampling fasta file during each training update. The UniRef50 training fasta was sorted in the order of occurrence of records in column 1 of the mapping file. The UniRef90+UniRef50 sampling fasta file was sorted in the order of occurrence of records in column 2 of the mapping file. Protein sequences longer than 1024 amino acids were cropped to 1022 from sequence start [3].
Unlike ESM-2 pre-training data, the curated pre-training dataset provided with ESM-2nv release contains hits for de novo proteins, since sequences in UniRef100, UniRef90, and UniRef50 with high sequence similarity to a non-public 81 de novo proteins [1] are not filtered.
Evaluation Dataset:#
Link: FLIP – secondary structure, conservation,subcellular localization, meltome, GB1 activity
Data Collection Method by dataset
[Human]
[Automatic/Sensors]
Labeling Method by dataset
[Experimentally Measured]
[Hybrid: Human & Automated]
Properties: The FLIP datasets evaluate the performance of the model on five specific downstream tasks for proteins. It provides pre-defined splits for fine-tuning a pretrained model using task-specific train and validation examples, and subsequently evaluating it on a task-specific test split.
The secondary structure FLIP dataset contains experimental secondary structures, with 9712 proteins for model finetuning, 1080 proteins for validation, and 648 proteins for testing.
Conservation dataset contains conservation scores of the residues of protein sequences with 9392 proteins for training, 555 proteins for validation, and 519 proteins for testing.
Subcellular localization dataset contains protein subcellular locations with 9503 proteins for training, 1678 proteins for validation, and 2768 proteins for testing.
Meltome dataset contains experimental melting temperatures for proteins, with 22335 proteins for training, 2482 proteins for validation, and 3134 proteins for testing.
The GB1 activity dataset contains experimental binding affinities of GB1 protein variants with variation at four sites (V39, D40, G41 and V54) measured in a binding assay, with 6289 proteins for training, 699 proteins for validation, and 1745 proteins for testing.
License Data:
Dataset License(s): AFL-3
Inference:#
Engine: BioNeMo, NeMo
Test Hardware:
[Ampere]
[Hopper]
[Volta]
Accuracy Benchmarks#
The accuracy of ESM2-nv was measured using the Fitness Landscape Inference for Proteins (FLIP) benchmarks [Dallago et al., 2022], which involve training a downstream task on top of the ESM2-nv model and measuring some characteristic:
FLIP type |
Dataset |
Dataset Split for Measurement |
Metric |
650m |
3b |
|---|---|---|---|---|---|
Secondary Structure |
sequences.fasta |
test |
accuracy (%) |
85.2 |
85.7 |
GB1 |
two_vs_rest.fasta |
test |
rmse (score) |
1.29 |
1.28 |
Residue Conservation |
sequences.fasta |
test |
accuracy (%) |
33.2 |
34.5 |
Protein Melting Point (Meltome) |
mixed_split.fasta |
test |
rmse (°C) |
7.19 |
6.68 |
Training Performance Benchmarks#
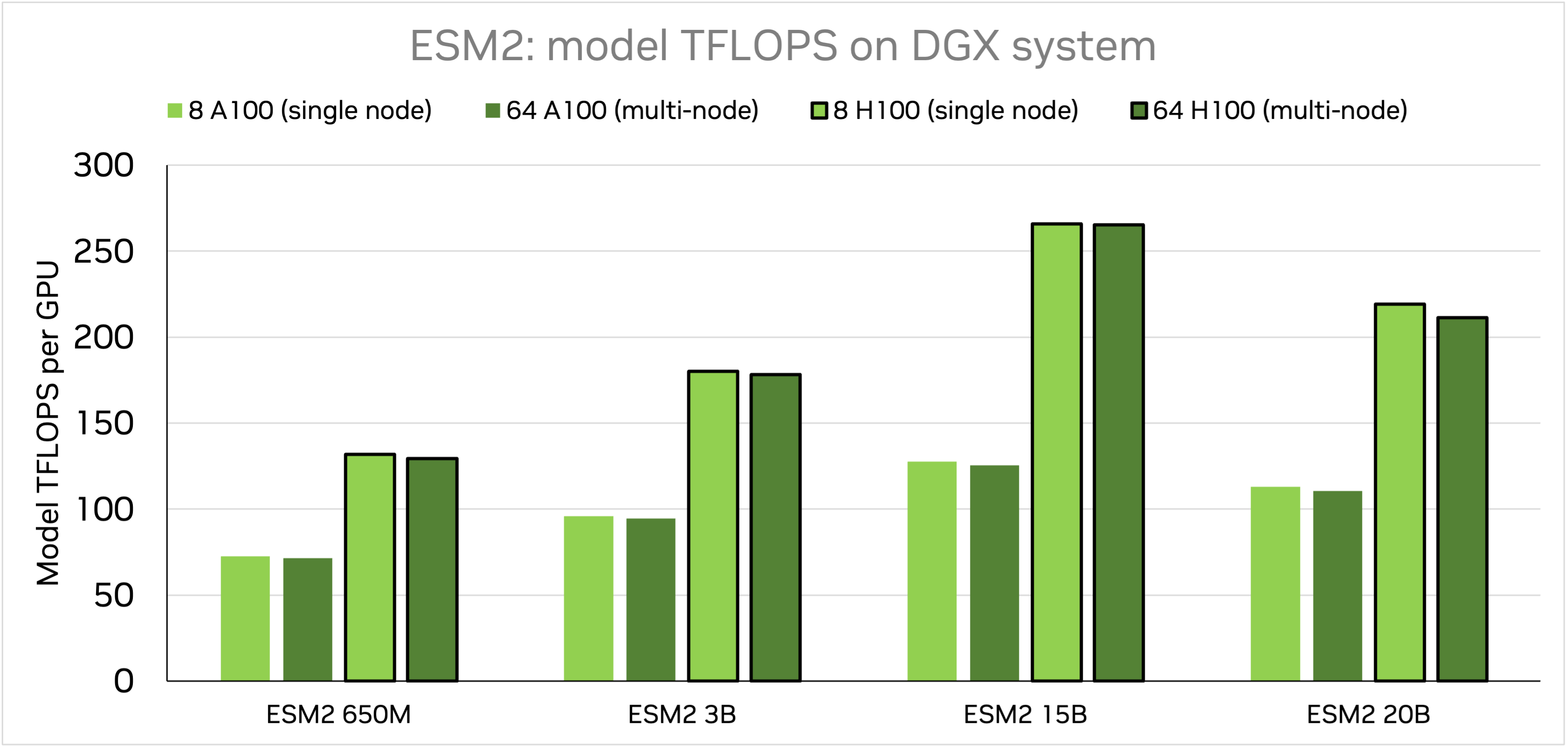
Training speed was tested on DGX-A100 and DGX-H100 systems, on GPUs with 80GB of memory.
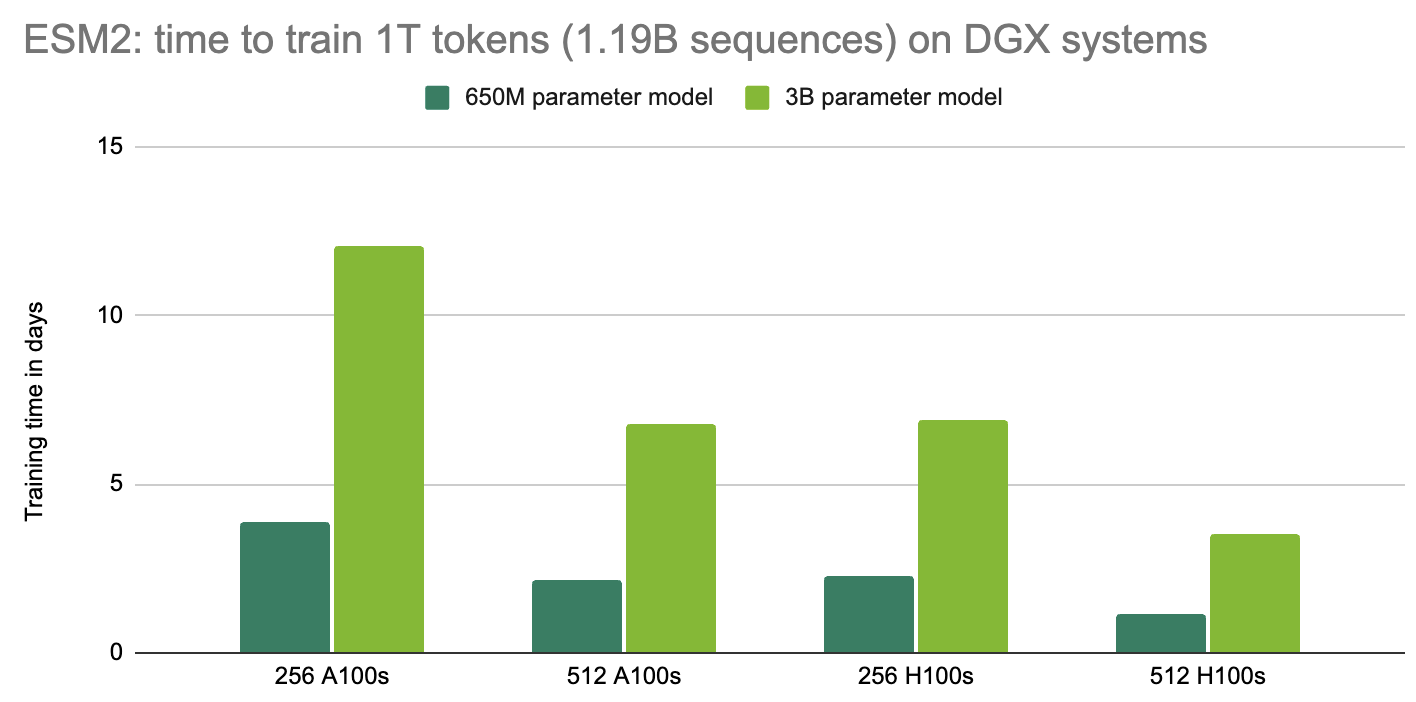
Model TFLOPS were collected for trainings of different model sizes of ESM-2nv on DGX-A100 and DGX-H100. Two comparisions were made: 1) single-node training using 1 node (8 GPUs), and 2) multi-node training using 8 nodes (64 GPUs). While the implementation of EMS2 in BioNeMo FW makes use of dynamic padding, we keep fixed sequence length of 1024. All models were trained using tensor model parallel of 1 and accumulated gradient of 32, but batch sizes and pipeline model parallel depend on model size:
ESM-2nv 650M: 650M parameter model, batch size of 16, pipeline parallel 1
ESM-2nv 3B: 3B parameters model, batch size of 4, pipeline parallel 4
ESM-2nv 15B: 15B parameter model, batch size of 2, pipeline parallel 8
ESM-2nv 20B: 20B parameters model, batch size of 1, pipeline parallel 8
License#
ESM-2nv is as provided under the Apache License.
