MolMIM Property Guided Molecular Optimization Using CMA-ES
Contents
MolMIM Property Guided Molecular Optimization Using CMA-ES#
Here we demonstrate how to load a MolMIM checkpoint from the BioNeMo Framework and use it to optimize some molecules of interest with a custom user-defined scoring function. We use CMA-ES to traverse the latent space of our MolMIM model and select novel, related molecules expected to improve performance as measured by the scoring function. To sample these molecules, we must complete the following steps:
Load the desired MolMIM checkpoint.
Encode the starting molecules into MolMIM’s latent space.
Run CMA-ES, which will iteratively perform the following:
Decode latent representations into SMILES strings.
Apply the user defined scoring function to these SMILES strings to generate SMILES/scores pairings.
Ask the CMA-ES algorithm for a new set of latent space representations from which to sample.
Note: this notebook is derived from a previous tutorial made for the BioNeMo Service version of MolMIM.
Setup your environment for this test#
For this tutorial, we assume you are running within the latest BioNeMo Framework Docker container.
From within the Docker container, download the example checkpoint, or use your own:
python download_models.py --download_dir models molmim_70m_24_3
Load your checkpoint into the molmim inference wrapper#
from bionemo.utils.hydra import load_model_config
import os
from bionemo.model.molecule.molmim.infer import MolMIMInference
bionemo_home=f"/workspace/bionemo"
os.environ['BIONEMO_HOME'] = bionemo_home
checkpoint_path = f"{bionemo_home}/models/molecule/molmim/molmim_70m_24_3.nemo"
cfg = load_model_config(config_name="molmim_infer.yaml", config_path=f"{bionemo_home}/examples/tests/conf/") # reasonable starting config for molmim inference
# This is the field of the config that we need to set to our desired checkpoint path.
cfg.model.downstream_task.restore_from_path = checkpoint_path
model = MolMIMInference(cfg, interactive=True)
Show code cell output
[NeMo I 2024-03-22 15:50:47 megatron_hiddens:110] Registered hidden transform sampled_var_cond_gaussian at bionemo.model.core.hiddens_support.SampledVarGaussianHiddenTransform
[NeMo I 2024-03-22 15:50:47 megatron_hiddens:110] Registered hidden transform interp_var_cond_gaussian at bionemo.model.core.hiddens_support.InterpVarGaussianHiddenTransform
[NeMo I 2024-03-22 15:50:47 utils:326] Restoring model from /workspace/bionemo/models/molecule/molmim/molmim_70m_24_3.nemo
[NeMo I 2024-03-22 15:50:47 utils:330] Loading model class: bionemo.model.molecule.molmim.molmim_model.MolMIMModel
Interactive mode selected, using strategy='auto'
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
[NeMo I 2024-03-22 15:50:48 exp_manager:394] Experiments will be logged at /workspace/bionemo/test_results/nemo_experiments/molmim_infer/MolMIM_Inference/2024-03-22_15-50-48
[NeMo I 2024-03-22 15:50:48 utils:299]
************** Trainer configuration ***********
[NeMo I 2024-03-22 15:50:48 utils:300]
name: MolMIM_Inference
desc: Minimum configuration for initializing a MolMIM model for inference.
trainer:
precision: 16-mixed
devices: 1
num_nodes: 1
accelerator: gpu
logger: false
accumulate_grad_batches: 1
exp_manager:
explicit_log_dir: null
exp_dir: ${oc.env:BIONEMO_HOME}/test_results/nemo_experiments/molmim_infer
name: ${name}
create_checkpoint_callback: false
create_wandb_logger: false
create_tensorboard_logger: false
wandb_logger_kwargs:
offline: true
model:
encoder:
num_layers: 6
hidden_size: 512
ffn_hidden_size: 2048
num_attention_heads: 8
init_method_std: 0.02
hidden_dropout: 0.1
attention_dropout: 0.1
ffn_dropout: 0.0
position_embedding_type: learned_absolute
relative_attention_num_buckets: 32
relative_attention_max_distance: 128
relative_position_bias_self_attention_only: true
kv_channels: null
apply_query_key_layer_scaling: false
layernorm_epsilon: 1.0e-05
persist_layer_norm: true
bias_activation_fusion: true
grad_div_ar_fusion: true
masked_softmax_fusion: true
bias_dropout_add_fusion: true
bias: true
normalization: layernorm
arch: perceiver
activation: gelu
headscale: false
transformer_block_type: pre_ln
hidden_steps: 1
num_self_attention_per_cross_attention: 1
openai_gelu: false
onnx_safe: false
fp32_residual_connection: false
activations_checkpoint_method: null
activations_checkpoint_num_layers: 1
activations_checkpoint_granularity: null
megatron_legacy: false
normalize_attention_scores: true
num_moe_experts: 1
moe_frequency: 1
moe_dropout: 0.0
use_flash_attention: false
decoder:
num_layers: 6
hidden_size: 512
ffn_hidden_size: 2048
num_attention_heads: 8
init_method_std: 0.02
hidden_dropout: 0.1
attention_dropout: 0.1
ffn_dropout: 0.0
position_embedding_type: learned_absolute
relative_attention_num_buckets: 32
relative_attention_max_distance: 128
relative_position_bias_self_attention_only: true
kv_channels: null
apply_query_key_layer_scaling: false
layernorm_epsilon: 1.0e-05
persist_layer_norm: true
bias_activation_fusion: true
grad_div_ar_fusion: true
masked_softmax_fusion: true
bias_dropout_add_fusion: true
bias: true
normalization: layernorm
arch: transformer
activation: gelu
headscale: false
transformer_block_type: pre_ln
hidden_steps: 32
num_self_attention_per_cross_attention: 1
openai_gelu: false
onnx_safe: false
fp32_residual_connection: false
activations_checkpoint_method: null
activations_checkpoint_num_layers: 1
activations_checkpoint_granularity: null
megatron_legacy: false
normalize_attention_scores: true
num_moe_experts: 1
moe_frequency: 1
moe_dropout: 0.0
use_flash_attention: false
name: MolMIM-small
micro_batch_size: ${model.data.batch_size}
global_batch_size: 128
tensor_model_parallel_size: 1
pipeline_model_parallel_size: 1
resume_from_checkpoint: null
pipeline_model_parallel_split_rank: 0
make_vocab_size_divisible_by: 128
pre_process: true
post_process: true
megatron_amp_O2: false
seq_length: 128
max_position_embeddings: 128
gradient_as_bucket_view: true
bias_gelu_fusion: true
share_token_embeddings: true
share_decoder_tokens_head_embeddings: false
hidden_size: 512
training_callbacks: []
hiddens:
enc_output_name: z
enc_inference_output_name: z_mean
token_aggregation_method: mean
hidden_aggregation_method: mean
transform:
q_z_given_x:
cls_name: sampled_var_cond_gaussian
hidden_size: 512
min_logvar: -6.0
max_logvar: 0.0
map_var_to_hiddens: false
loss:
mim:
cls_name: a_mim
loss_weight: 1.0
tokenizer:
library: regex
type: null
model: nemo:048c1f797f464dd5b6a90f60f9405827_molmim.model
vocab_file: nemo:dd344353154640acbbaea1d4536fa7d0_molmim.vocab
merge_file: null
vocab_path: ${oc.env:BIONEMO_HOME}/tokenizers/molecule/molmim/vocab/molmim.vocab
model_path: ${oc.env:BIONEMO_HOME}/tokenizers/molecule/molmim/vocab/molmim.model
data:
links_file: /workspace/bionemo/examples/molecule/megamolbart/dataset/ZINC-downloader.txt
dataset_path: ${oc.env:BIONEMO_HOME}/examples/tests/test_data/molecule/physchem/SAMPL/test/x000
dataset:
train: x_OP_000..175_CL_
test: x_OP_000..175_CL_
val: x_OP_000..004_CL_
canonicalize_target_smile: true
canonicalize_encoder_input: true
canonicalize_decoder_output: true
encoder_augment: false
decoder_independent_augment: false
encoder_mask: false
decoder_mask: false
mask_prob: 0.0
span_lambda: 3.0
micro_batch_size: 2048
num_workers: 4
dataloader_type: single
max_seq_length: 128
seed: 42
skip_lines: 0
drop_last: false
pin_memory: false
data_impl: ''
index_mapping_type: online
data_impl_kwargs:
csv_mmap:
newline_int: 10
header_lines: 1
workers: 10
sort_dataset_paths: true
data_sep: ','
data_col: 1
csv_fields_mmap:
newline_int: 10
header_lines: 1
workers: null
sort_dataset_paths: false
data_sep: ','
data_fields:
id: 0
sequence: 1
fasta_fields_mmap:
data_fields:
id: 0
sequence: 1
use_upsampling: true
index_mapping_dir: null
batch_size: 128
output_fname: ${oc.env:BIONEMO_HOME}/test_results/nemo_experiments/molmim_infer/x000.pkl
data_fields_map:
sequence: smiles
id: iupac
optim:
name: fused_adam
lr: 0.0005
weight_decay: 0.001
betas:
- 0.9
- 0.999
sched:
name: CosineAnnealing
warmup_steps: 10000.0
constant_steps: 50000.0
max_steps: 1000000
min_lr: 5.0e-05
dwnstr_task_validation:
enabled: false
dataset:
class: bionemo.model.core.dwnstr_task_callbacks.SingleValuePredictionCallback
task_type: regression
infer_target: bionemo.model.molecule.molmim.infer.MolMIMInference
max_seq_length: 128
emb_batch_size: 128
batch_size: 128
num_epochs: 10
shuffle: true
num_workers: 8
dataset_path: /data/physchem/
task_name: SAMPL
dataset:
train: x000
test: x000
sequence_column: smiles
target_column: expt
random_seed: 1234
optim:
name: adam
lr: 0.0001
betas:
- 0.9
- 0.999
eps: 1.0e-08
weight_decay: 0.01
sched:
name: WarmupAnnealing
min_lr: 1.0e-05
last_epoch: -1
warmup_ratio: 0.01
max_steps: 1000
precision: 32
target: bionemo.model.molecule.molmim.molmim_model.MolMIMModel
nemo_version: 1.22.0
downstream_task:
restore_from_path: /workspace/bionemo/models/molecule/molmim/molmim_70m_24_3.nemo
outputs:
- embeddings
target: bionemo.model.molecule.molmim.molmim_model.MolMIMModel
infer_target: bionemo.model.molecule.molmim.infer.MolMIMInference
formatters:
simple:
format: '[%(asctime)s][%(name)s][%(levelname)s] - %(message)s'
handlers:
console:
class: logging.StreamHandler
formatter: simple
stream: ext://sys.stdout
file:
class: logging.FileHandler
formatter: simple
filename: /logs/inference.log
root:
level: INFO
handlers:
- console
disable_existing_loggers: false
infer_config:
name: MolMIM_Inference
desc: Store the infer config in this block so we can pull the model path from it
later.
trainer:
precision: 16-mixed
devices: 1
num_nodes: 1
accelerator: gpu
logger: false
exp_manager:
explicit_log_dir: null
exp_dir: null
name: ${name}
create_checkpoint_callback: false
model:
micro_batch_size: ${model.data.batch_size}
downstream_task:
restore_from_path: ${oc.env:BIONEMO_HOME}/models/molecule/molmim/molmim_70m_24_3.nemo
outputs:
- embeddings
data:
num_workers: 4
batch_size: 128
dataset_path: ${oc.env:BIONEMO_HOME}/examples/tests/test_data/molecule/physchem/SAMPL/test/x000
output_fname: ''
index_mapping_dir: null
data_fields_map:
sequence: smiles
id: iupac
data_impl: ''
data_impl_kwargs:
csv_fields_mmap:
newline_int: 10
header_lines: 1
workers: null
sort_dataset_paths: false
data_sep: ','
data_fields:
id: 0
sequence: 1
fasta_fields_mmap:
data_fields:
id: 0
sequence: 1
training_callbacks: []
tokenizer:
vocab_path: ${oc.env:BIONEMO_HOME}/tokenizers/molecule/molmim/vocab/molmim.vocab
model_path: ${oc.env:BIONEMO_HOME}/tokenizers/molecule/molmim/vocab/molmim.model
target: bionemo.model.molecule.molmim.molmim_model.MolMIMModel
infer_target: bionemo.model.molecule.molmim.infer.MolMIMInference
formatters:
simple:
format: '[%(asctime)s][%(name)s][%(levelname)s] - %(message)s'
handlers:
console:
class: logging.StreamHandler
formatter: simple
stream: ext://sys.stdout
file:
class: logging.FileHandler
formatter: simple
filename: /logs/inference.log
root:
level: INFO
handlers:
- console
disable_existing_loggers: false
hydra:
searchpath:
- file://${oc.env:BIONEMO_HOME}/examples/conf/
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: context_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: virtual_pipeline_model_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: sequence_parallel in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: expert_model_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: use_cpu_initialization in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: gradient_accumulation_fusion in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_overlap in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_split_ag in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_split_rs in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_bulk_wgrad in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_bulk_dgrad in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: finalize_model_grads_func in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: overlap_p2p_comm in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: batch_p2p_comm in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: barrier_with_L1_time in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo I 2024-03-22 15:50:48 megatron_init:234] Rank 0 has data parallel group: [0]
[NeMo I 2024-03-22 15:50:48 megatron_init:237] All data parallel group ranks: [[0]]
[NeMo I 2024-03-22 15:50:48 megatron_init:238] Ranks 0 has data parallel rank: 0
[NeMo I 2024-03-22 15:50:48 megatron_init:246] Rank 0 has model parallel group: [0]
[NeMo I 2024-03-22 15:50:48 megatron_init:247] All model parallel group ranks: [[0]]
[NeMo I 2024-03-22 15:50:48 megatron_init:257] Rank 0 has tensor model parallel group: [0]
[NeMo I 2024-03-22 15:50:48 megatron_init:261] All tensor model parallel group ranks: [[0]]
[NeMo I 2024-03-22 15:50:48 megatron_init:262] Rank 0 has tensor model parallel rank: 0
[NeMo I 2024-03-22 15:50:48 megatron_init:276] Rank 0 has pipeline model parallel group: [0]
[NeMo I 2024-03-22 15:50:48 megatron_init:288] Rank 0 has embedding group: [0]
[NeMo I 2024-03-22 15:50:48 megatron_init:294] All pipeline model parallel group ranks: [[0]]
[NeMo I 2024-03-22 15:50:48 megatron_init:295] Rank 0 has pipeline model parallel rank 0
[NeMo I 2024-03-22 15:50:48 megatron_init:296] All embedding group ranks: [[0]]
[NeMo I 2024-03-22 15:50:48 megatron_init:297] Rank 0 has embedding rank: 0
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: context_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: virtual_pipeline_model_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: sequence_parallel in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: expert_model_parallel_size in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: use_cpu_initialization in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: gradient_accumulation_fusion in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_overlap in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_split_ag in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_split_rs in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_bulk_wgrad in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: tp_comm_bulk_dgrad in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: finalize_model_grads_func in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: overlap_p2p_comm in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: batch_p2p_comm in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 megatron_base_model:821] The model: MolMIMModel() does not have field.name: barrier_with_L1_time in its cfg. Add this key to cfg or config_mapping to make to make it configurable.
[NeMo W 2024-03-22 15:50:48 modelPT:251] You tried to register an artifact under config key=tokenizer.vocab_file but an artifact for it has already been registered.
[NeMo I 2024-03-22 15:50:48 tokenizer_utils:199] Using regex tokenization
[NeMo I 2024-03-22 15:50:48 regex_tokenizer:240] Loading vocabulary from file = /tmp/tmpkrp5q227/dd344353154640acbbaea1d4536fa7d0_molmim.vocab
[NeMo I 2024-03-22 15:50:48 regex_tokenizer:254] Loading regex from file = /tmp/tmpkrp5q227/048c1f797f464dd5b6a90f60f9405827_molmim.model
[NeMo I 2024-03-22 15:50:48 megatron_base_model:315] Padded vocab_size: 640, original vocab_size: 523, dummy tokens: 117.
[NeMo I 2024-03-22 15:50:48 megatron_hiddens:121] NOTE: Adding hiddens transforms and losses
[NeMo I 2024-03-22 15:50:48 megatron_hiddens:149] Added transform q_z_given_x with cfg={'cls_name': 'sampled_var_cond_gaussian', 'hidden_size': 512, 'min_logvar': -6.0, 'max_logvar': 0.0, 'map_var_to_hiddens': False}
[NeMo I 2024-03-22 15:50:48 megatron_hiddens:177] Added loss mim with cfg={'cls_name': 'a_mim', 'loss_weight': 1.0}
[NeMo I 2024-03-22 15:50:49 nlp_overrides:752] Model MolMIMModel was successfully restored from /workspace/bionemo/models/molecule/molmim/molmim_70m_24_3.nemo.
[NeMo W 2024-03-22 15:50:49 nemo_logging:349] /usr/local/lib/python3.10/dist-packages/nemo/collections/nlp/modules/common/megatron/fused_bias_dropout_add.py:70: UserWarning: nvfuser integration in TorchScript is deprecated. (Triggered internally at /opt/pytorch/pytorch/torch/csrc/jit/codegen/cuda/interface.cpp:235.)
return bias_dropout_add_fused_inference_(*args)
[NeMo I 2024-03-22 15:50:50 megatron_lm_encoder_decoder_model:1195] Decoding using the greedy-search method...
Setup user-defined molecule scoring function#
This is the section where you as a user can pull in your own scoring functions that you want to optimize. For this example, we will be optimizing a combination of Tanimoto similarity to the input molecule and Quantitative Estimate of Druglikeness (QED) following the example from the initial MolMIM publication:
In this case, we will allow the model to optimize up to a maximum QED of 0.9 and Tanimoto similarity of 0.4. Once these maxima are achieved, we perform no further optimization.
from typing import List, Optional
import numpy as np
from guided_molecule_gen.oracles import qed, tanimoto_similarity
def score_mixing_function(qeds, similarities):
# We want to maximize QED and tanimoto similarity up to 0.9 and 0.4, respectively.
return np.clip(qeds / 0.9, a_min=0.0, a_max=1.0) + np.clip(similarities / 0.4, a_min=0.0, a_max=1.0)
def try_canon(smiles:str) -> Optional[str]:
try:
return Chem.MolToSmiles(Chem.MolFromSmiles(smiles), canonical=True)
except:
return None
def canonicalize(smiles: List[str]) -> List[str]:
return [try_canon(s) for s in smiles]
def scoring_function(smiles: List[str], reference:str, **kwargs) -> np.ndarray:
"""Takes a list of SMILES strings and returns an array of scores.
Args:
smiles (List[str]): Smiles strings to generate a score for (one each)
reference (str): Reference molecule (SMILES string) is also used for this scoring function.
Returns:
np.ndarray: Array of scores, one for each input SMILES string.
"""
#csmiles = canonicalize(smiles)
scores: np.ndarray = score_mixing_function(qed(smiles), tanimoto_similarity(smiles, reference))
return -1 * scores
Define starting molecules#
In this section, we will define the starting molecules for the optimization process. As a set of examples, we will use imatinib, erlotinib, and gifitinib. We ensure that the SMILES strings representing these molecules are canonicalized using RDKit. MolMIM was trained on a corpus of RDKit-cononicalized SMILES strings, so any inputs and outputs should be RDKit-canonicalized as well to achieve peak performance.
from rdkit import Chem
from rdkit.Chem.QED import qed as rdkit_qed
starting_smiles = [
"CC1=C(C=C(C=C1)NC(=O)C2=CC=C(C=C2)CN3CCN(CC3)C)NC4=NC=CC(=N4)C5=CN=CC=C5", # imatinib
"COCCOC1=C(C=C2C(=C1)C(=NC=N2)NC3=CC=CC(=C3)C#C)OCCOC", # erlotinib
"C1COCCN1CCCOc2c(OC)cc3ncnc(c3c2)Nc4cc(Cl)c(F)cc4", # gifitinib
]
# Canonicalize all SMILES strings and print the structure of imatinib
molecules = [Chem.MolFromSmiles(s) for s in starting_smiles]
starting_qed = [rdkit_qed(m) for m in molecules]
canonicalized_smiles = [Chem.MolToSmiles(m, canonical=True) for m in molecules]
molecules[0]
Setup the optimizer and wrap the inference API for CMA-ES#
The CMA-ES library expects certain formats for input/output of the inference model to function properly. We provide a wrapper for this and show how to setup optimization below.
from bionemo.model.core.controlled_generation import ControlledGenerationPerceiverEncoderInferenceWrapper
controlled_gen_kwargs = {
"sampling_method": "beam-search",
"sampling_kwarg_overrides": {"beam_size": 3, "keep_only_best_tokens": True, "return_scores": False},
}
model_wrapped = ControlledGenerationPerceiverEncoderInferenceWrapper(
model, enforce_perceiver=True, hidden_steps=1, **controlled_gen_kwargs
) # just flatten the position for this.
Tune CMA-ES#
Different models will have different optimal settings for CMA-ES. Here, we perform a grid search over possible values of sigma, then perform more steps of optimization with the best. We will use the Optuna library to perform this optimization over the sigma hyperparameter. This process is referred to as hyperparatemer optimization or HPO.
!pip install optuna
Show code cell output
Defaulting to user installation because normal site-packages is not writeable
Looking in indexes: https://pypi.org/simple, https://pypi.ngc.nvidia.com
Requirement already satisfied: optuna in /workspace/bionemo/.local/lib/python3.10/site-packages (3.5.0)
Requirement already satisfied: alembic>=1.5.0 in /workspace/bionemo/.local/lib/python3.10/site-packages (from optuna) (1.13.1)
Requirement already satisfied: colorlog in /workspace/bionemo/.local/lib/python3.10/site-packages (from optuna) (6.8.2)
Requirement already satisfied: numpy in /usr/local/lib/python3.10/dist-packages (from optuna) (1.23.5)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.10/dist-packages (from optuna) (23.1)
Requirement already satisfied: sqlalchemy>=1.3.0 in /workspace/bionemo/.local/lib/python3.10/site-packages (from optuna) (2.0.28)
Requirement already satisfied: tqdm in /usr/local/lib/python3.10/dist-packages (from optuna) (4.66.1)
Requirement already satisfied: PyYAML in /usr/local/lib/python3.10/dist-packages (from optuna) (6.0.1)
Requirement already satisfied: Mako in /workspace/bionemo/.local/lib/python3.10/site-packages (from alembic>=1.5.0->optuna) (1.3.2)
Requirement already satisfied: typing-extensions>=4 in /usr/local/lib/python3.10/dist-packages (from alembic>=1.5.0->optuna) (4.7.1)
Requirement already satisfied: greenlet!=0.4.17 in /usr/local/lib/python3.10/dist-packages (from sqlalchemy>=1.3.0->optuna) (3.0.3)
Requirement already satisfied: MarkupSafe>=0.9.2 in /usr/local/lib/python3.10/dist-packages (from Mako->alembic>=1.5.0->optuna) (2.1.3)
[notice] A new release of pip is available: 23.2.1 -> 24.0
[notice] To update, run: python -m pip install --upgrade pip
from guided_molecule_gen.optimizer import MoleculeGenerationOptimizer
import optuna
def objective(trial, n_steps:int=10):
sigma = trial.suggest_float('sigma', 0, 2)
optimizer = MoleculeGenerationOptimizer(
model_wrapped,
scoring_function,
canonicalized_smiles,
popsize=10, # larger values will be slower but more thorough
optimizer_args={"sigma": sigma},
)
optimizer.optimize(n_steps)
final_smiles = optimizer.generated_smis
final_score = np.mean([np.min(scoring_function(smis_population, reference_smis)) for smis_population,reference_smis in zip(final_smiles, canonicalized_smiles)])
return final_score
study = optuna.create_study()
study.optimize(objective, n_trials=50)
print(study.best_params)
Show code cell output
[I 2024-03-22 15:50:56,086] A new study created in memory with name: no-name-7e3f6666-25c5-4af0-a8a3-987ea1477566
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=184028, Fri Mar 22 15:50:56 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=263743, Fri Mar 22 15:50:56 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=177481, Fri Mar 22 15:50:56 2024)
[NeMo I 2024-03-22 15:50:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:50:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:00 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:51:15,771] Trial 0 finished with value: -1.6638290331236025 and parameters: {'sigma': 0.13176216745842084}. Best is trial 0 with value: -1.6638290331236025.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=268975, Fri Mar 22 15:51:15 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=213670, Fri Mar 22 15:51:15 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=251428, Fri Mar 22 15:51:15 2024)
[NeMo I 2024-03-22 15:51:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:51:34,970] Trial 1 finished with value: -1.6708456377350098 and parameters: {'sigma': 0.1788389080326782}. Best is trial 1 with value: -1.6708456377350098.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=302939, Fri Mar 22 15:51:34 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=131562, Fri Mar 22 15:51:35 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=153099, Fri Mar 22 15:51:35 2024)
[NeMo I 2024-03-22 15:51:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:51:54,100] Trial 2 finished with value: -0.9968631161177796 and parameters: {'sigma': 1.7341544339881114}. Best is trial 1 with value: -1.6708456377350098.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=159782, Fri Mar 22 15:51:54 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=215392, Fri Mar 22 15:51:54 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=213395, Fri Mar 22 15:51:54 2024)
[NeMo I 2024-03-22 15:51:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:51:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:52:13,295] Trial 3 finished with value: -1.4509472129725 and parameters: {'sigma': 1.5698095134424892}. Best is trial 1 with value: -1.6708456377350098.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=228388, Fri Mar 22 15:52:13 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=223598, Fri Mar 22 15:52:13 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=194880, Fri Mar 22 15:52:13 2024)
[NeMo I 2024-03-22 15:52:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:52:32,490] Trial 4 finished with value: -1.6573820968029167 and parameters: {'sigma': 1.3099029637878306}. Best is trial 1 with value: -1.6708456377350098.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=206956, Fri Mar 22 15:52:32 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=260273, Fri Mar 22 15:52:32 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=203825, Fri Mar 22 15:52:32 2024)
[NeMo I 2024-03-22 15:52:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:52:51,741] Trial 5 finished with value: -1.7754158666744182 and parameters: {'sigma': 0.35769460248621776}. Best is trial 5 with value: -1.7754158666744182.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=231723, Fri Mar 22 15:52:51 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=206547, Fri Mar 22 15:52:51 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=255305, Fri Mar 22 15:52:51 2024)
[NeMo I 2024-03-22 15:52:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:52:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:53:10,863] Trial 6 finished with value: -1.337132995342649 and parameters: {'sigma': 1.957306121905405}. Best is trial 5 with value: -1.7754158666744182.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=257384, Fri Mar 22 15:53:10 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=283855, Fri Mar 22 15:53:10 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=275855, Fri Mar 22 15:53:10 2024)
[NeMo I 2024-03-22 15:53:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:53:30,091] Trial 7 finished with value: -1.8120665871264265 and parameters: {'sigma': 1.0946556916301828}. Best is trial 7 with value: -1.8120665871264265.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=144259, Fri Mar 22 15:53:30 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=206699, Fri Mar 22 15:53:30 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=180830, Fri Mar 22 15:53:30 2024)
[NeMo I 2024-03-22 15:53:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:37 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:39 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:53:49,369] Trial 8 finished with value: -1.7738668287052797 and parameters: {'sigma': 1.316710963004739}. Best is trial 7 with value: -1.8120665871264265.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=212574, Fri Mar 22 15:53:49 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=240296, Fri Mar 22 15:53:49 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=166781, Fri Mar 22 15:53:49 2024)
[NeMo I 2024-03-22 15:53:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:53:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:54:08,784] Trial 9 finished with value: -1.8039128660064192 and parameters: {'sigma': 0.28291842967817105}. Best is trial 7 with value: -1.8120665871264265.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=275187, Fri Mar 22 15:54:08 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=232382, Fri Mar 22 15:54:08 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=229828, Fri Mar 22 15:54:08 2024)
[NeMo I 2024-03-22 15:54:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:54:28,177] Trial 10 finished with value: -1.926194020904745 and parameters: {'sigma': 0.7517222599082064}. Best is trial 10 with value: -1.926194020904745.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=163545, Fri Mar 22 15:54:28 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=168658, Fri Mar 22 15:54:28 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=169644, Fri Mar 22 15:54:28 2024)
[NeMo I 2024-03-22 15:54:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:37 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:39 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:54:47,311] Trial 11 finished with value: -1.9423986990758306 and parameters: {'sigma': 0.7532818500298163}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=201897, Fri Mar 22 15:54:47 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=180255, Fri Mar 22 15:54:47 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=224534, Fri Mar 22 15:54:47 2024)
[NeMo I 2024-03-22 15:54:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:54:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:00 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:55:06,489] Trial 12 finished with value: -1.9047630129013011 and parameters: {'sigma': 0.64025838012875}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=223828, Fri Mar 22 15:55:06 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=255712, Fri Mar 22 15:55:06 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=217592, Fri Mar 22 15:55:06 2024)
[NeMo I 2024-03-22 15:55:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:55:25,663] Trial 13 finished with value: -1.8963051721567767 and parameters: {'sigma': 0.6853723356903602}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=280846, Fri Mar 22 15:55:25 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=292116, Fri Mar 22 15:55:25 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=199572, Fri Mar 22 15:55:25 2024)
[NeMo I 2024-03-22 15:55:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:37 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:39 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:55:44,797] Trial 14 finished with value: -1.9186069296744457 and parameters: {'sigma': 0.7544391124205039}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=252994, Fri Mar 22 15:55:44 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=294518, Fri Mar 22 15:55:44 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=255612, Fri Mar 22 15:55:44 2024)
[NeMo I 2024-03-22 15:55:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:55:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:00 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:56:03,999] Trial 15 finished with value: -1.7499775285813346 and parameters: {'sigma': 0.9405113283810009}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=223726, Fri Mar 22 15:56:04 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=147863, Fri Mar 22 15:56:04 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=145696, Fri Mar 22 15:56:04 2024)
[NeMo I 2024-03-22 15:56:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:56:23,359] Trial 16 finished with value: -1.8721884918273846 and parameters: {'sigma': 0.5555891150803465}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=235632, Fri Mar 22 15:56:23 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=260134, Fri Mar 22 15:56:23 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=184070, Fri Mar 22 15:56:23 2024)
[NeMo I 2024-03-22 15:56:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:56:42,600] Trial 17 finished with value: -1.9057377513153446 and parameters: {'sigma': 0.9964055132222357}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=246718, Fri Mar 22 15:56:42 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=231716, Fri Mar 22 15:56:42 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=232281, Fri Mar 22 15:56:42 2024)
[NeMo I 2024-03-22 15:56:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:56:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:57:01,846] Trial 18 finished with value: -1.9092861368330676 and parameters: {'sigma': 0.4551612841742564}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=251906, Fri Mar 22 15:57:01 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=231697, Fri Mar 22 15:57:01 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=264823, Fri Mar 22 15:57:01 2024)
[NeMo I 2024-03-22 15:57:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:57:21,143] Trial 19 finished with value: -1.8626183161109882 and parameters: {'sigma': 0.844995506528921}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=239436, Fri Mar 22 15:57:21 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=173264, Fri Mar 22 15:57:21 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=215070, Fri Mar 22 15:57:21 2024)
[NeMo I 2024-03-22 15:57:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:57:40,386] Trial 20 finished with value: -1.9324065421120828 and parameters: {'sigma': 1.2310733125891054}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=194599, Fri Mar 22 15:57:40 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=186390, Fri Mar 22 15:57:40 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=174718, Fri Mar 22 15:57:40 2024)
[NeMo I 2024-03-22 15:57:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:57:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:57:59,677] Trial 21 finished with value: -1.8428655891494188 and parameters: {'sigma': 1.1818364561163708}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=252500, Fri Mar 22 15:57:59 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=290590, Fri Mar 22 15:57:59 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=286948, Fri Mar 22 15:57:59 2024)
[NeMo I 2024-03-22 15:57:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:58:18,963] Trial 22 finished with value: -1.6811500963907349 and parameters: {'sigma': 1.5308198469573666}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=226541, Fri Mar 22 15:58:18 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=322521, Fri Mar 22 15:58:18 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=186856, Fri Mar 22 15:58:19 2024)
[NeMo I 2024-03-22 15:58:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:58:38,217] Trial 23 finished with value: -1.8941596701526364 and parameters: {'sigma': 0.8233070618875522}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=241831, Fri Mar 22 15:58:38 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=222120, Fri Mar 22 15:58:38 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=205221, Fri Mar 22 15:58:38 2024)
[NeMo I 2024-03-22 15:58:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:58:57,463] Trial 24 finished with value: -1.8815083468261118 and parameters: {'sigma': 1.268977937364232}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=202703, Fri Mar 22 15:58:57 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=195518, Fri Mar 22 15:58:57 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=195087, Fri Mar 22 15:58:57 2024)
[NeMo I 2024-03-22 15:58:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:58:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:59:16,723] Trial 25 finished with value: -1.6182401109767266 and parameters: {'sigma': 0.9429778755439713}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=199634, Fri Mar 22 15:59:16 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=271269, Fri Mar 22 15:59:16 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=286306, Fri Mar 22 15:59:16 2024)
[NeMo I 2024-03-22 15:59:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:59:36,055] Trial 26 finished with value: -1.5085159203262488 and parameters: {'sigma': 0.0151811479211722}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=194872, Fri Mar 22 15:59:36 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=135098, Fri Mar 22 15:59:36 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=188585, Fri Mar 22 15:59:36 2024)
[NeMo I 2024-03-22 15:59:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:39 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 15:59:55,286] Trial 27 finished with value: -1.4522338288887677 and parameters: {'sigma': 1.4496218566771268}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=213882, Fri Mar 22 15:59:55 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=192385, Fri Mar 22 15:59:55 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=228897, Fri Mar 22 15:59:55 2024)
[NeMo I 2024-03-22 15:59:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 15:59:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:00:14,637] Trial 28 finished with value: -1.8963760110895127 and parameters: {'sigma': 0.4976237739915497}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=214946, Fri Mar 22 16:00:14 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=241094, Fri Mar 22 16:00:14 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=258835, Fri Mar 22 16:00:14 2024)
[NeMo I 2024-03-22 16:00:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:00:33,708] Trial 29 finished with value: -1.8242226396025376 and parameters: {'sigma': 1.0451019644408983}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=285065, Fri Mar 22 16:00:33 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=245849, Fri Mar 22 16:00:33 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=273138, Fri Mar 22 16:00:33 2024)
[NeMo I 2024-03-22 16:00:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:37 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:39 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:00:52,819] Trial 30 finished with value: -1.9218879026458857 and parameters: {'sigma': 0.6515837398976043}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=289490, Fri Mar 22 16:00:52 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=275849, Fri Mar 22 16:00:52 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=273881, Fri Mar 22 16:00:52 2024)
[NeMo I 2024-03-22 16:00:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:00:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:00 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:01:12,012] Trial 31 finished with value: -1.9338713394264195 and parameters: {'sigma': 0.6485299757678998}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=140200, Fri Mar 22 16:01:12 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=156825, Fri Mar 22 16:01:12 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=157927, Fri Mar 22 16:01:12 2024)
[NeMo I 2024-03-22 16:01:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:01:31,136] Trial 32 finished with value: -1.9333597782225072 and parameters: {'sigma': 0.8444994761777667}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=179666, Fri Mar 22 16:01:31 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=178777, Fri Mar 22 16:01:31 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=218185, Fri Mar 22 16:01:31 2024)
[NeMo I 2024-03-22 16:01:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:01:50,311] Trial 33 finished with value: -1.7757320324091639 and parameters: {'sigma': 1.1293370664926266}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=234626, Fri Mar 22 16:01:50 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=249918, Fri Mar 22 16:01:50 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=230717, Fri Mar 22 16:01:50 2024)
[NeMo I 2024-03-22 16:01:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:01:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:02:09,465] Trial 34 finished with value: -1.902726188737371 and parameters: {'sigma': 0.8808139308833965}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=225156, Fri Mar 22 16:02:09 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=238235, Fri Mar 22 16:02:09 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=200335, Fri Mar 22 16:02:09 2024)
[NeMo I 2024-03-22 16:02:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:02:28,688] Trial 35 finished with value: -1.7620145937657286 and parameters: {'sigma': 0.30869355372841156}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=208857, Fri Mar 22 16:02:28 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=278194, Fri Mar 22 16:02:28 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=220499, Fri Mar 22 16:02:28 2024)
[NeMo I 2024-03-22 16:02:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:02:47,762] Trial 36 finished with value: -1.4791671475133459 and parameters: {'sigma': 1.743253379935838}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=216354, Fri Mar 22 16:02:47 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=264587, Fri Mar 22 16:02:47 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=299831, Fri Mar 22 16:02:47 2024)
[NeMo I 2024-03-22 16:02:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:02:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:03:06,945] Trial 37 finished with value: -1.8893909740584895 and parameters: {'sigma': 0.566589151399223}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=255357, Fri Mar 22 16:03:06 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=312641, Fri Mar 22 16:03:06 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=284156, Fri Mar 22 16:03:06 2024)
[NeMo I 2024-03-22 16:03:06 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:08 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:12 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:14 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:03:26,363] Trial 38 finished with value: -1.755344061231452 and parameters: {'sigma': 0.38099996961078}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=173160, Fri Mar 22 16:03:26 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=190173, Fri Mar 22 16:03:26 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=179125, Fri Mar 22 16:03:26 2024)
[NeMo I 2024-03-22 16:03:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:43 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:03:45,884] Trial 39 finished with value: -1.5279037949916094 and parameters: {'sigma': 1.2469911815050108}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=246384, Fri Mar 22 16:03:45 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=258923, Fri Mar 22 16:03:45 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=262125, Fri Mar 22 16:03:45 2024)
[NeMo I 2024-03-22 16:03:45 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:49 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:51 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:03:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:04:05,360] Trial 40 finished with value: -1.4979116048202694 and parameters: {'sigma': 1.4425849699281745}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=204855, Fri Mar 22 16:04:05 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=175613, Fri Mar 22 16:04:05 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=243517, Fri Mar 22 16:04:05 2024)
[NeMo I 2024-03-22 16:04:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:18 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:20 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:04:24,787] Trial 41 finished with value: -1.914962596062159 and parameters: {'sigma': 0.7790948272215702}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=207243, Fri Mar 22 16:04:24 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=302593, Fri Mar 22 16:04:24 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=250671, Fri Mar 22 16:04:24 2024)
[NeMo I 2024-03-22 16:04:24 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:26 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:30 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:04:44,315] Trial 42 finished with value: -1.8984086192947387 and parameters: {'sigma': 0.7435943516156961}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=179932, Fri Mar 22 16:04:44 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=250011, Fri Mar 22 16:04:44 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=162163, Fri Mar 22 16:04:44 2024)
[NeMo I 2024-03-22 16:04:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:55 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:04:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:01 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:05:03,755] Trial 43 finished with value: -1.930160329186462 and parameters: {'sigma': 0.5938764880213937}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=260548, Fri Mar 22 16:05:03 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=249059, Fri Mar 22 16:05:03 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=203210, Fri Mar 22 16:05:03 2024)
[NeMo I 2024-03-22 16:05:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:05:23,370] Trial 44 finished with value: -1.6666490543092618 and parameters: {'sigma': 0.17483916895703122}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=225759, Fri Mar 22 16:05:23 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=196378, Fri Mar 22 16:05:23 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=189967, Fri Mar 22 16:05:23 2024)
[NeMo I 2024-03-22 16:05:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:33 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:05:42,820] Trial 45 finished with value: -1.8265102544770533 and parameters: {'sigma': 0.6180491725804795}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=267685, Fri Mar 22 16:05:42 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=295700, Fri Mar 22 16:05:42 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=215375, Fri Mar 22 16:05:42 2024)
[NeMo I 2024-03-22 16:05:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:05:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:00 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:06:01,974] Trial 46 finished with value: -1.8795102727263056 and parameters: {'sigma': 0.4134911520364928}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=136209, Fri Mar 22 16:06:02 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=200756, Fri Mar 22 16:06:02 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=214650, Fri Mar 22 16:06:02 2024)
[NeMo I 2024-03-22 16:06:02 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:03 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:07 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:09 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:13 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:15 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:19 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:06:21,291] Trial 47 finished with value: -1.8341060706918597 and parameters: {'sigma': 0.944221123662994}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=163991, Fri Mar 22 16:06:21 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=202888, Fri Mar 22 16:06:21 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=201267, Fri Mar 22 16:06:21 2024)
[NeMo I 2024-03-22 16:06:21 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:25 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:27 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:31 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:32 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:36 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:38 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:06:40,601] Trial 48 finished with value: -1.8977260716725437 and parameters: {'sigma': 0.5307976908704173}. Best is trial 11 with value: -1.9423986990758306.
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=209752, Fri Mar 22 16:06:40 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=274104, Fri Mar 22 16:06:40 2024)
(5_w,10)-aCMA-ES (mu_w=3.2,w_1=45%) in dimension 512 (seed=220422, Fri Mar 22 16:06:40 2024)
[NeMo I 2024-03-22 16:06:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:42 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:44 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:48 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:50 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:54 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:56 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[NeMo I 2024-03-22 16:06:57 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
[I 2024-03-22 16:06:59,969] Trial 49 finished with value: -1.9471215994488233 and parameters: {'sigma': 0.7181389906909391}. Best is trial 49 with value: -1.9471215994488233.
{'sigma': 0.7181389906909391}
Now, we can examine the best performing value for sigma found by Optuna during HPO.
study.best_params
{'sigma': 0.7181389906909391}
Though the above value is the optimium returned over our HPO process, we will consider the range of valid values and pick a minimum that is more likely to be robust. Since the HPO process is stochastic, high-performing and low-performing values may be in close proximity. We would like to identify a good range of sigma values, over which the optimizer generally performs well.
!pip install statsmodels
Defaulting to user installation because normal site-packages is not writeable
Looking in indexes: https://pypi.org/simple, https://pypi.ngc.nvidia.com
Requirement already satisfied: statsmodels in /workspace/bionemo/.local/lib/python3.10/site-packages (0.14.1)
Requirement already satisfied: numpy<2,>=1.18 in /usr/local/lib/python3.10/dist-packages (from statsmodels) (1.23.5)
Requirement already satisfied: scipy!=1.9.2,>=1.4 in /usr/local/lib/python3.10/dist-packages (from statsmodels) (1.11.1)
Requirement already satisfied: pandas!=2.1.0,>=1.0 in /usr/local/lib/python3.10/dist-packages (from statsmodels) (1.5.3)
Requirement already satisfied: patsy>=0.5.4 in /workspace/bionemo/.local/lib/python3.10/site-packages (from statsmodels) (0.5.6)
Requirement already satisfied: packaging>=21.3 in /usr/local/lib/python3.10/dist-packages (from statsmodels) (23.1)
Requirement already satisfied: python-dateutil>=2.8.1 in /usr/local/lib/python3.10/dist-packages (from pandas!=2.1.0,>=1.0->statsmodels) (2.8.2)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.10/dist-packages (from pandas!=2.1.0,>=1.0->statsmodels) (2023.3)
Requirement already satisfied: six in /usr/local/lib/python3.10/dist-packages (from patsy>=0.5.4->statsmodels) (1.16.0)
[notice] A new release of pip is available: 23.2.1 -> 24.0
[notice] To update, run: python -m pip install --upgrade pip
import statsmodels.api as sm
import pandas as pd
completed_trials = [trial for trial in study.trials if trial.state == optuna.trial.TrialState.COMPLETE]
trials_data = [{"sigma": trial.params["sigma"], "loss": trial.value, "trial_id": tid} for tid,trial in enumerate(completed_trials)]
data = pd.DataFrame(trials_data)
# Now create a bootstrap confidence interval around the a LOWESS fit
def lowess_with_confidence_bounds(
x, y, eval_x, N=200, conf_interval=0.95, lowess_kw=None
):
"""
Perform Lowess regression and determine a confidence interval by bootstrap resampling
"""
# Lowess smoothing
# x is called (exog), y is called (endog)
smoothed = sm.nonparametric.lowess(exog=x, endog=y, xvals=eval_x, **lowess_kw)
# Perform bootstrap resamplings of the data
# and evaluate the smoothing at a fixed set of points
smoothed_values = np.empty((N, len(eval_x)))
for i in range(N):
sample = np.random.choice(len(x), len(x), replace=True)
sampled_x = x[sample]
sampled_y = y[sample]
smoothed_values[i] = sm.nonparametric.lowess(
exog=sampled_x, endog=sampled_y, xvals=eval_x, **lowess_kw
)
# Get the confidence interval
sorted_values = np.sort(smoothed_values, axis=0)
bound = int(N * (1 - conf_interval) / 2)
bottom = sorted_values[bound - 1]
top = sorted_values[-bound]
return smoothed, bottom, top
# Compute the 95% confidence interval
eval_x = np.linspace(0, 2, 200)
smoothed, bottom, top = lowess_with_confidence_bounds(
data.sigma, data.loss, eval_x, lowess_kw={"frac": 0.33}
)
import matplotlib.pyplot as plt
plt.scatter(x=data["sigma"], y=data["loss"], label="observed_points")
plt.plot(eval_x, smoothed, c="k", label="lowess smoothed")
plt.fill_between(eval_x, bottom, top, alpha=0.5, color="b", label="lowess 95% CI")
plt.legend()
plt.title("Loss vs sigma from HPO for MolMIM model")
plt.autoscale(enable=True, axis="x", tight=True);
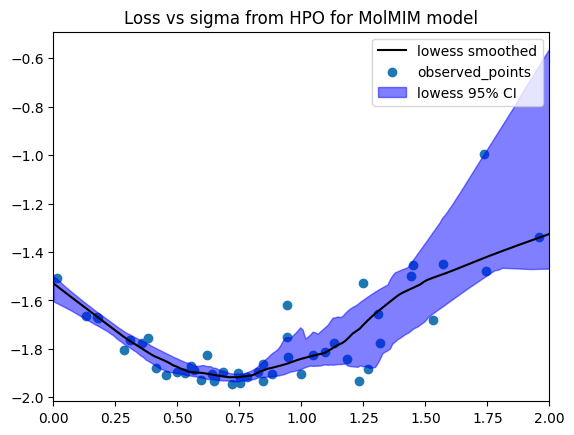
smoothed_best_sigma = eval_x[np.argmin(top)] # Use the upper bound of the confidence interval
smooth_best = {"sigma": smoothed_best_sigma}
smooth_best
{'sigma': 0.7437185929648241}
We now compare the smoothed top choice with the best nominal choice.
smooth_best, study.best_params
({'sigma': 0.7437185929648241}, {'sigma': 0.7181389906909391})
Run a larger CMA-ES optimization with discovered parameters#
Given the value of sigma we found to work well in our HPO above, we will increase the population size and steps and do a final larger optimizaiton run.
from tqdm import trange
optimizer = MoleculeGenerationOptimizer(
model_wrapped,
scoring_function,
canonicalized_smiles,
popsize=50, # larger values will be slower but more thorough
optimizer_args=smooth_best, # Vals from HPO
)
# Starting state for idx 0
qed_scores = [qed(canonicalized_smiles)]
tanimoto_scores = [[tanimoto_similarity([canonicalized_smiles[idx]], canonicalized_smiles[idx])[0] for idx in range(len(canonicalized_smiles))]]
best_molecules = [canonicalized_smiles]
fraction_bad_samples = [[0]*len(canonicalized_smiles)]
for i in trange(30):
optimizer.step()
final_smiles = optimizer.generated_smis
# Population of molecules is returned, but we only want the best one.
_qed_scores = []
_tanimoto_scores = []
_best_molecules = []
_fraction_bad = []
for smis_population,reference_smis in zip(final_smiles, canonicalized_smiles):
idx = np.argmin(scoring_function(smis_population, reference_smis))
_fraction_bad.append(np.mean(qed(smis_population) == 0))
_best_molecules.append(smis_population[idx])
_qed_scores.append(qed([smis_population[idx]])[0])
_tanimoto_scores.append(tanimoto_similarity([smis_population[idx]], reference_smis)[0])
qed_scores.append(_qed_scores)
tanimoto_scores.append(_tanimoto_scores)
best_molecules.append(_best_molecules)
fraction_bad_samples.append(_fraction_bad)
Show code cell output
(25_w,50)-aCMA-ES (mu_w=14.0,w_1=14%) in dimension 512 (seed=212874, Fri Mar 22 16:11:40 2024)
(25_w,50)-aCMA-ES (mu_w=14.0,w_1=14%) in dimension 512 (seed=270731, Fri Mar 22 16:11:40 2024)
(25_w,50)-aCMA-ES (mu_w=14.0,w_1=14%) in dimension 512 (seed=239948, Fri Mar 22 16:11:40 2024)
0%| | 0/30 [00:00<?, ?it/s]
[NeMo I 2024-03-22 16:11:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
3%|▎ | 1/30 [00:05<02:51, 5.93s/it]
[NeMo I 2024-03-22 16:11:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
7%|▋ | 2/30 [00:11<02:45, 5.92s/it]
[NeMo I 2024-03-22 16:11:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
10%|█ | 3/30 [00:17<02:41, 5.98s/it]
[NeMo I 2024-03-22 16:11:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
13%|█▎ | 4/30 [00:23<02:34, 5.95s/it]
[NeMo I 2024-03-22 16:12:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
17%|█▋ | 5/30 [00:29<02:29, 5.99s/it]
[NeMo I 2024-03-22 16:12:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
20%|██ | 6/30 [00:35<02:23, 5.96s/it]
[NeMo I 2024-03-22 16:12:16 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
23%|██▎ | 7/30 [00:41<02:19, 6.04s/it]
[NeMo I 2024-03-22 16:12:22 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
27%|██▋ | 8/30 [00:47<02:12, 6.02s/it]
[NeMo I 2024-03-22 16:12:28 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
30%|███ | 9/30 [00:54<02:06, 6.04s/it]
[NeMo I 2024-03-22 16:12:34 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
33%|███▎ | 10/30 [01:00<02:00, 6.02s/it]
[NeMo I 2024-03-22 16:12:40 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
37%|███▋ | 11/30 [01:06<01:54, 6.05s/it]
[NeMo I 2024-03-22 16:12:46 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
40%|████ | 12/30 [01:12<01:48, 6.03s/it]
[NeMo I 2024-03-22 16:12:52 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
43%|████▎ | 13/30 [01:18<01:43, 6.06s/it]
[NeMo I 2024-03-22 16:12:58 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
47%|████▋ | 14/30 [01:24<01:36, 6.04s/it]
[NeMo I 2024-03-22 16:13:04 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
50%|█████ | 15/30 [01:30<01:30, 6.07s/it]
[NeMo I 2024-03-22 16:13:10 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
53%|█████▎ | 16/30 [01:36<01:24, 6.05s/it]
[NeMo I 2024-03-22 16:13:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
57%|█████▋ | 17/30 [01:42<01:18, 6.08s/it]
[NeMo I 2024-03-22 16:13:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
60%|██████ | 18/30 [01:48<01:12, 6.06s/it]
[NeMo I 2024-03-22 16:13:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
63%|██████▎ | 19/30 [01:54<01:06, 6.08s/it]
[NeMo I 2024-03-22 16:13:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
67%|██████▋ | 20/30 [02:00<01:00, 6.06s/it]
[NeMo I 2024-03-22 16:13:41 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
70%|███████ | 21/30 [02:06<00:54, 6.07s/it]
[NeMo I 2024-03-22 16:13:47 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
73%|███████▎ | 22/30 [02:12<00:48, 6.05s/it]
[NeMo I 2024-03-22 16:13:53 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
77%|███████▋ | 23/30 [02:18<00:42, 6.08s/it]
[NeMo I 2024-03-22 16:13:59 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
80%|████████ | 24/30 [02:24<00:36, 6.05s/it]
[NeMo I 2024-03-22 16:14:05 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
83%|████████▎ | 25/30 [02:31<00:30, 6.07s/it]
[NeMo I 2024-03-22 16:14:11 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
87%|████████▋ | 26/30 [02:37<00:24, 6.05s/it]
[NeMo I 2024-03-22 16:14:17 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
90%|█████████ | 27/30 [02:43<00:18, 6.07s/it]
[NeMo I 2024-03-22 16:14:23 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
93%|█████████▎| 28/30 [02:49<00:12, 6.05s/it]
[NeMo I 2024-03-22 16:14:29 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
97%|█████████▋| 29/30 [02:55<00:06, 6.08s/it]
[NeMo I 2024-03-22 16:14:35 megatron_lm_encoder_decoder_model:1192] Decoding using the beam search method with beam size=3...
100%|██████████| 30/30 [03:01<00:00, 6.04s/it]
Explore results#
Below, we create a plot disaplaying how the components of our target (QED and Tanimoto similarity) changed over each iteration. By our target definition, any value above 0.4 for Tanimoto similarity would be optimal so we expect noise around that value. Similarly, for QED any value above 0.9 would be optimal so we expect noise around that value if any molecule surpasses that threshold.
import matplotlib.pyplot as plt
for i, molecule in enumerate(["imatinib", "erlotinib", "gifitinib"]):
line, = plt.plot(np.arange(len(qed_scores)), [q[i] for q in qed_scores], label=f"{molecule} QED")
color = line.get_color()
plt.plot(np.arange(len(tanimoto_scores)), [t[i] for t in tanimoto_scores], label=f"{molecule} Tanimoto", linestyle="--", color=color)
plt.axhline(y=0.9, color='r', linestyle='-', label="QED target")
plt.axhline(y=0.4, color='r', linestyle='--', label="Tanimoto target")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.xlabel("Iteration")
plt.ylabel("QED or Tanimoto similarity")
plt.title("Targets over time for MolMIM model")
Text(0.5, 1.0, 'Targets over time for MolMIM model')
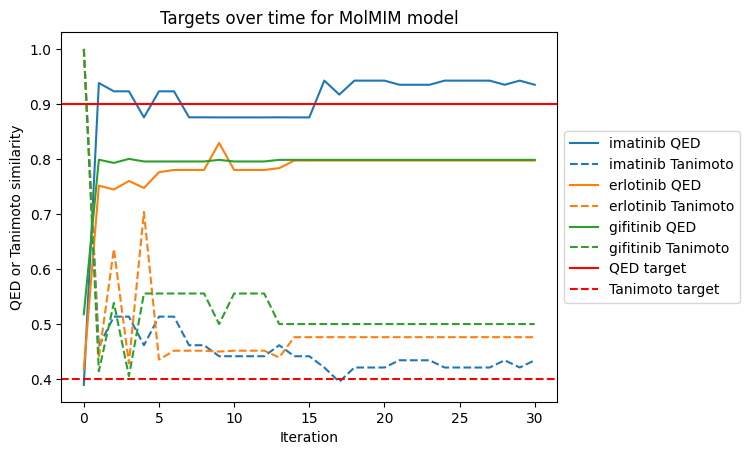
How well did our optimization perform?#
To examine the performance of out optimization, we can quantify the number of invalid samples that were generated. An “invalid” SMILES is defined as a SMILES string that does not represent a chemically-valid underlying molecule.
np.mean(fraction_bad_samples)
0.0015053763440860217
We can finally quantify the improvement in QED over the baseline value and the fraction of our optimized molecules that maintained the desired Tanimoto similarity threshold above 0.4.
qed_improvements = []
tanimoto_above_04 = []
for i in range(len(starting_qed)):
tanimoto_above_04.append(tanimoto_scores[-1][i] >= 0.4)
qed_improvements.append(qed_scores[-1][i] - starting_qed[i])
{"mean_qed_improvement": np.mean(qed_improvements), "tanimoto_above_04": np.mean(tanimoto_above_04)}
{'mean_qed_improvement': 0.40173431938851917, 'tanimoto_above_04': 1.0}
