ESM1nv Model Training using BioNeMo
Contents
ESM1nv Model Training using BioNeMo#
The purpose of this tutorial is to provide an example use case of training a BioNeMo Large Language model using the BioNeMo framework. At the end of this tutorial, the user will get experience in
configuring various config files and launch parameters for ESM-1nv training
launching single and multi-node, multi-gpu training runs
using NVIDIA’s Base Command Platform commands for LLM model training
Note
This tutorial focuses on ESM-1nv model training as an example, and the walk through can be easily modified for performing ProotT5-nv model training. The relevant config files and scripts for ProtT5-nv are provided in /workspace/bionemo/examples/protein/prott5nv/.
Overview - ESM1nv model#
ESM-1nv is based on the BERT architecture and trained on millions of protein sequences from the UniProt database. ESM-1nv learns the patterns and dependencies between amino acids that ultimately give rise to a protein’s 2D structure. These can include properties such as alpha helix or beta sheet, plus cellular location, thermostability, solubility, and other protein properties.
This ESM-1nv model training example walkthrough will show how to utilize the compute resources, download and preprocess the datasets, and perform model training on single and multiple nodes.
Setup and Assumptions#
This tutorial assumes that the user has access to BioNeMo framework and NVIDIA’s BCP and DGX-Cloud compute infrastructure. The user is also expected to have required background details about
the BioNeMo framework, as described in the Quickstart Guide, and
running the model training jobs on BCP
All model training related commands should be executed inside the BioNeMo docker container.
Requesting compute resources#
Access to DGX compute resource NGC site or NGC CLI#
As a prerequisite, configure your access to the DGX compute resources and required contents either via NVIDIA’s Base Command Platform or NGC-CLI using ngc config set command.
For more details on how to request the resources, visit Running BioNeMo on DGX-Cloud using BCP
Note
The interactive job launch example shown here is using interactive shell interface. It is strongly advised to launch the model training jobs using the launch script as a part of the ngc batch run command, as mentioned in Running BioNeMo on DGX-Cloud using BCP. For ESM1nv training, the model training script should be used as a template for launching the job as provided in <BioNeMO_Workspace>/examples/protein/esm1nv/scripts/pretrain_bcp_prd11.sh.
First, let’s request the resource for running the model training in an interactive manner.
Here is one such example of a command for requesting the resources using NGC-CLI. Make sure to update the relevant arguments according to the compute setup, datasets, workspaces, instance types, and so on.
In the configuration below, update nvidia and clara with the correct NGC org and team name, respectively. If there is no team name, then this can be omitted. Refer to NGC documentation for more details.
ngc batch run \
--name "example-training-1" \
--org nvidia \
--team clara \
--instance INSTANCE_TYPE \ #Compute node type, such as dgxa100.80g.8.norm
--array-type PYTORCH \
--replicas 2 \
--image "nvidia/clara/bionemo-framework:1.3" \ #Image path for BioNeMo
--result /results \
--workspace WORKSPACE_ID:/example_training:RW \
--port 8888 \
--datasetid DATASET_ID:/data/ \ # Dataset's NGC ID
--total-runtime 1D \
--preempt RUNONCE \
--priority MEDIUM \ # Priority level for the jog execution [LOW, MEDIUM, HIGH]
--order 1 \ # Priority order for the jog execution [1-99]
--commandline "sleep infinity" # This command can be replaced with the model training command: python pretrain.py....
The bcprun command provided in the cells below can also be submitted as --commandline argument (instead of launching interactive shell).
Once the resources are assigned for the job and the BioNeMo container is running, we’ll proceed ahead via ngc attach <JOB_ID>.
Data Preprocessing#
Downloading and pre-processing the dataset#
Download the data#
The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and annotation data [1].
The UniRef is a set of Reference Clusters with sequences from the UniProt Knowledge base and selected UniParc records. UniRef50 is a “second derivation” of UniRef100: Uniref90 is generated by clustering UniRef100 seed sequences and UniRef50 is generated by clustering UniRef90 sequences. For more information refer to the UniRef page.
Using BioNeMo features to download UniRef50#
The simplest and most reliable way to download the entire UniRef50 dataset is through the BioNeMo framework UniRef50Preprocess class which has the following features:
Runs a fasta indexer
Splits the data into train, validation and test samples
Writes the dataset in the appropriate directories within the BioNeMo Framework
/tmp/uniref50/processed
For example, here is the python code snippet for downloading and preprocessing the UniRef50 dataset.
from bionemo.data import UniRef50Preprocess
data = UniRef50Preprocess()
data.prepare_dataset()
In the snippet above, the UniRef50 (clusters) will be downloaded. However, for this example, we’ll pass the UniProtKB dataset as argument to the function above.
Alternative datasets#
We can also download datasets that are not available in the BioNeMo Framework. This can be done in two ways:
A) Using bash and wget pointing to the dataset’s URL
mkdir -p /tmp/data/protein/esm1nv
wget -P /tmp/data/protein/esm1nv <URL>
B) Transfering from the local machine to the container
docker cp <dataset directory and filename> container_id:/<container directory and filename>
Then, once the data is downloaded, we can start moving files and using the Data Loaders and Data Module to make sure the dataset is in a format the BioNeMo Framework can operate. It is not guaranteed that the UniRef50Preprocess class will handle datasets other than those from UniProt.
Model training#
Example dataset#
To briefly showcase the model training capacities of BioNeMo Framework, we will use a very small subset of the original UniRef50 dataset that is provided as a part of the sample datasets located in ${BIONEMO_HOME}/examples/tests/test_data/protein
For the purpose of this test run, the folder contains /train, /val, /test folders with protein sequences in CSV files.
Single-node or Multi-node setup#
In this test runs, we will use preconfigured parameters provided in the pretrain_small.yaml config file located in the ${BIONEMO_HOME}/examples/protein/esm1nv/conf folder.
We will also set other parameters suitable for a quick run, such as ++trainer.max_steps=100 or by using very limited protein subset as x000.csv file. User can update these parameters by editing the .yaml config file or as additional command line arguments, as shown in the example below. User can select the full dataset and adjust other parameters - for example - as shown in the base_config.yaml file.
As we are connected to the compute node, we navigate to the BioNeMo home folder using the command cd ${BIONEMO_HOME}, and execute the following command in the terminal.
User may need to update relevant arguments in the commands according to their compute and data setup.
Note
To run the model training job on a local workstation, user can directly execute the pretrain.py script with desired configurations. For example,
python examples/protein/esm1nv/pretrain.py
The bcprun command is similar to srun command in SLURM, you can find more details at the NVIDIA BCP User Guide.
Please make changes in the example command provided below as needed before running them.
bcprun --nnodes=1 --npernode=8 \
--cmd "python examples/protein/esm1nv/pretrain.py \
--config-path=conf \
--config-name=pretrain_small do_training=True model.data.dataset_path=examples/tests/test_data/protein \
++model.data.dataset.train=x000 ++model.data.dataset.val=x000 ++model.data.dataset.test=x000 ++exp_manager.wandb_logger_kwargs.offline=False \
++trainer.devices=8 ++trainer.num_nodes=1 ++model.validation.validation_enabled=False model.micro_batch_size=128 ++trainer.max_steps=100 \
++trainer.val_check_interval=12 ++exp_manager.create_wandb_logger=False ++model.tensor_model_parallel_size=1 \
++trainer.accumulate_grad_batches=1 ++exp_manager.checkpoint_callback_params.always_save_nemo=False \
++model.dwnstr_task_validation.dataset.dataset_path=examples/tests/test_data/protein/downstream trainer.precision=16-mixed"
To run the model training on multiple nodes, you will have to update parameters accordingly, for example, the command running the model training job on 4 nodes would look like:
bcprun --nnodes=4 --npernode=8 \
--cmd "python examples/protein/esm1nv/pretrain.py \
--config-path=conf \
--config-name=pretrain_small do_training=True model.data.dataset_path=examples/tests/test_data/protein \
++model.data.dataset.train=x000 ++model.data.dataset.val=x000 ++model.data.dataset.test=x000 ++exp_manager.wandb_logger_kwargs.offline=False \
++trainer.devices=8 ++trainer.num_nodes=4 ++model.validation.validation_enabled=False model.micro_batch_size=128 ++trainer.max_steps=100 \
++trainer.val_check_interval=12 ++exp_manager.create_wandb_logger=False ++model.tensor_model_parallel_size=1 \
++trainer.accumulate_grad_batches=1 ++exp_manager.checkpoint_callback_params.always_save_nemo=False \
++model.dwnstr_task_validation.dataset.dataset_path=examples/tests/test_data/protein/downstream trainer.precision=16-mixed"
Logging with WandB#
If you are launching the model training job interactively from the terminal, you can set your Weights and Biases access via wandb login <YOUR_WANDB_API_KEY> or checkout https://docs.wandb.ai/ref/cli/wandb-login for more information. Alternatively, you may also export the API key as a variable at the time of launching the job via command-line, as shown in ${BIONEMO_HOME}/examples/protein/esm1nv/scripts/pretrain_bcp_prd11.sh
Output and Results#
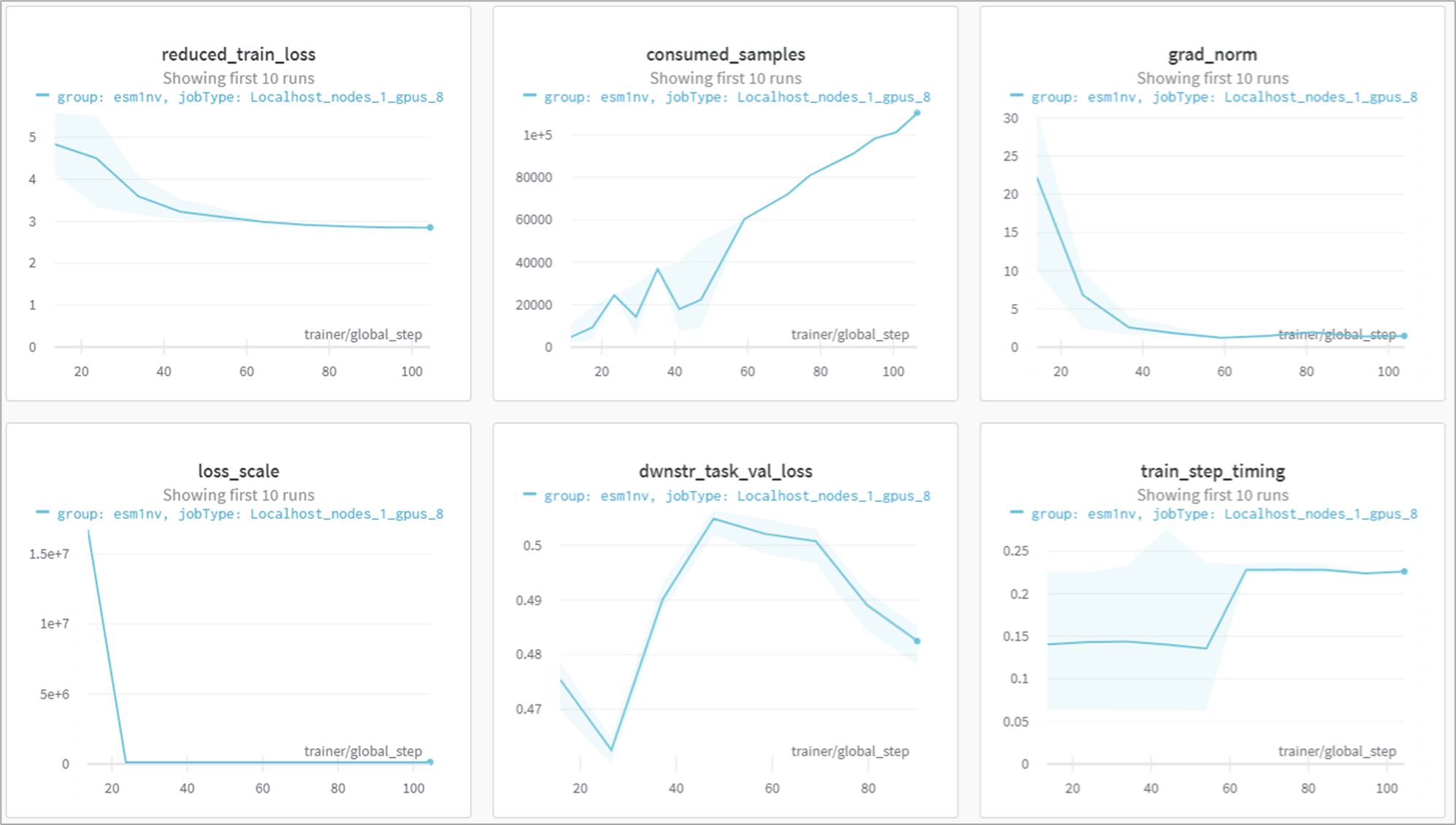
As the ESM1nv model training job is launched, BioNeMo will print out some of the details related to compute resources, model training configuration, and dataset being used for training. As the job progresses, it will also print out various details related to the test/train/validation steps and accuracy matrices at a set intervals.
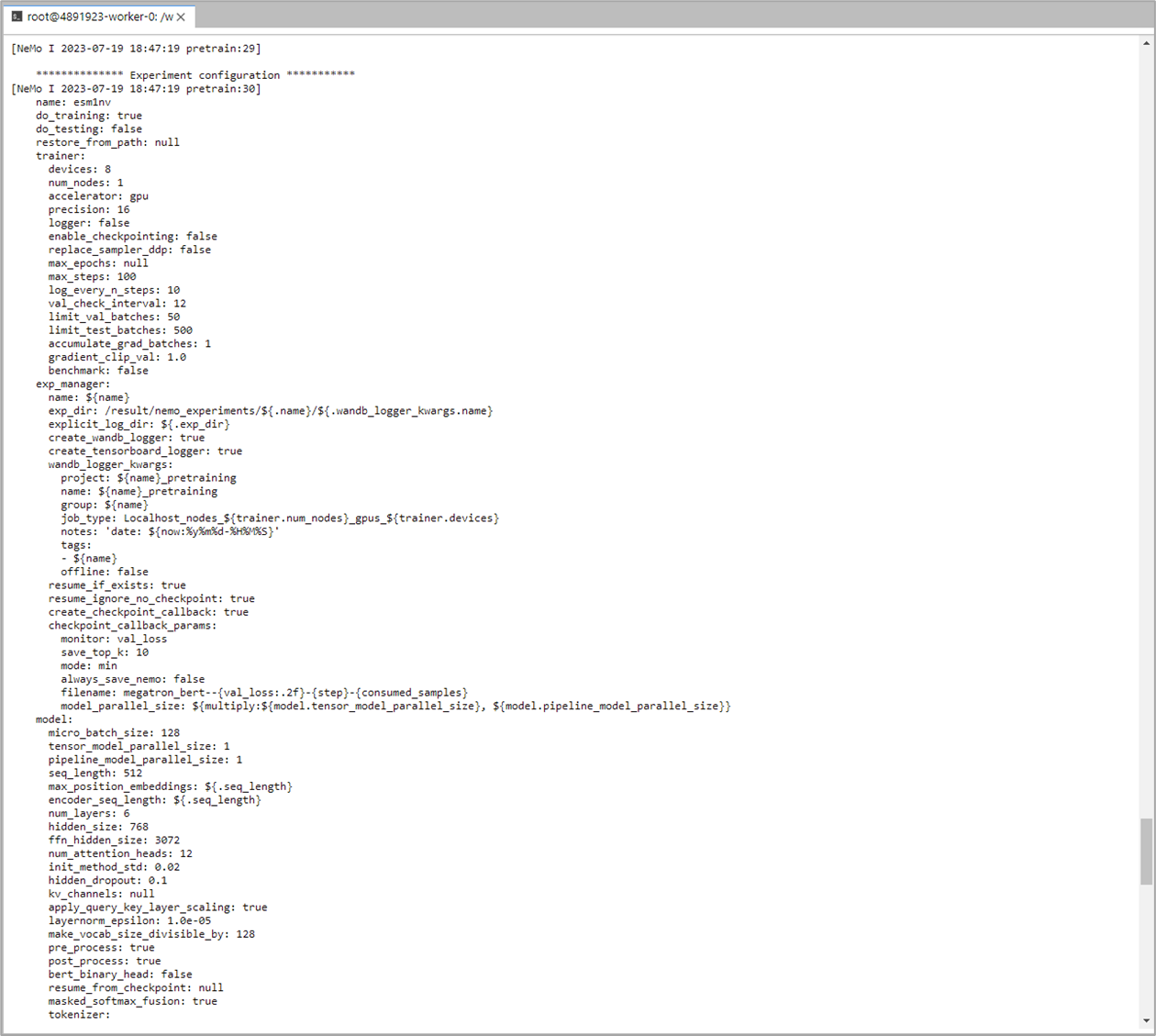
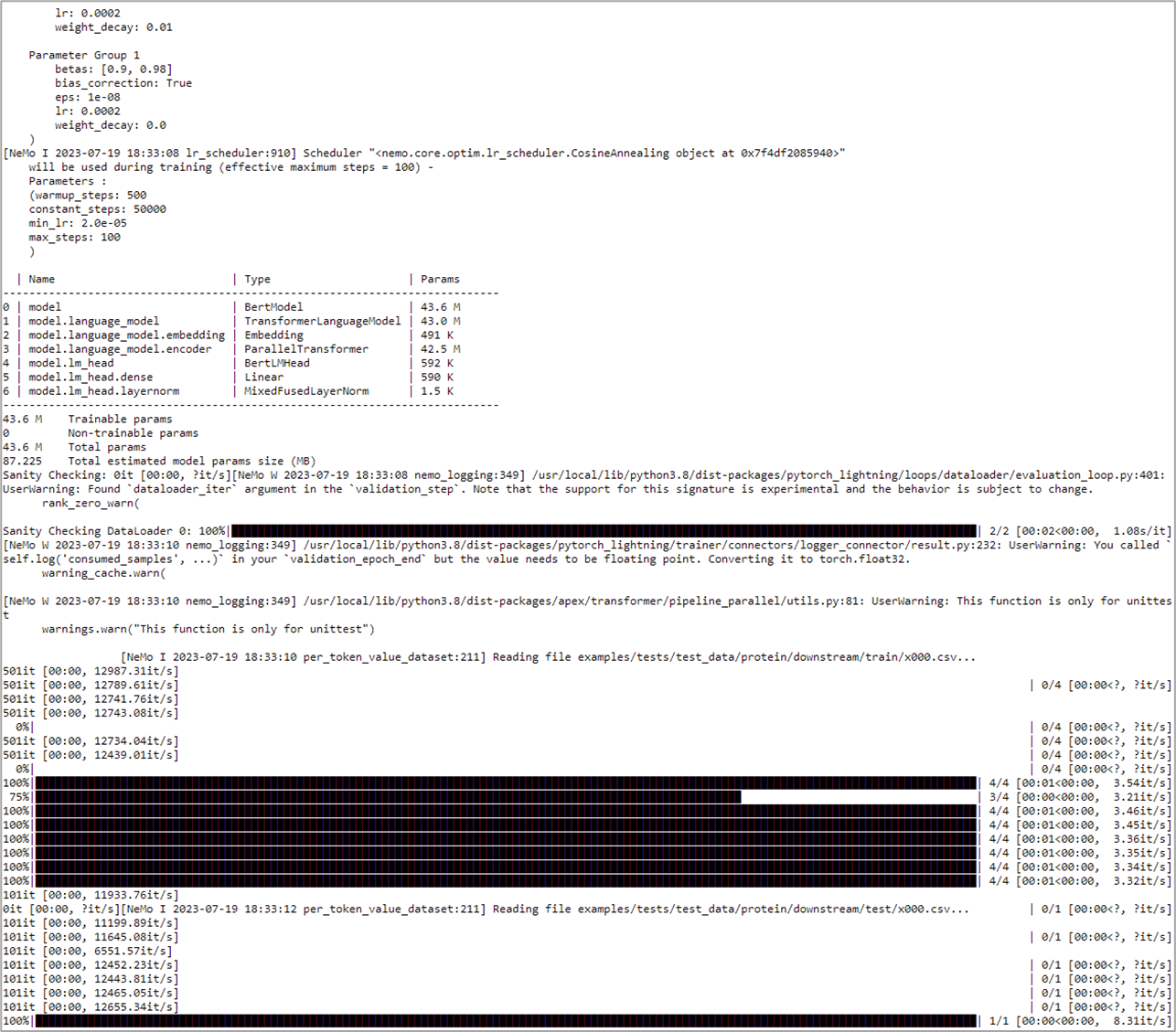
Upon the completion of training process, it will also print out the details related to log files, model checkpoints, and so on, that will also be saved in the directory as configured (usually /result).

Finally, if Weights and Biases logging was enabled (for example, ++exp_manager.create_wandb_logger=True ), you can also visualize the model training progress and resulting matrices, and the summary also gets printed on the termainal at the end of the training job.