Step #2: Setting Up a MONAI Label Slicer Module
Once the environment and the server up and running, we move to the viewer side. Inside the Desktop is the 3D Slicer software that you can use to interact with the MONAI Label module.
Skip the following steps if you’ve already started Slicer and installed MONAI Label for the Radiology App. There’s no need to install it again. Please go to the step Connecting to the MONAI Label server.
The Desktop can be found on the top left on this page:
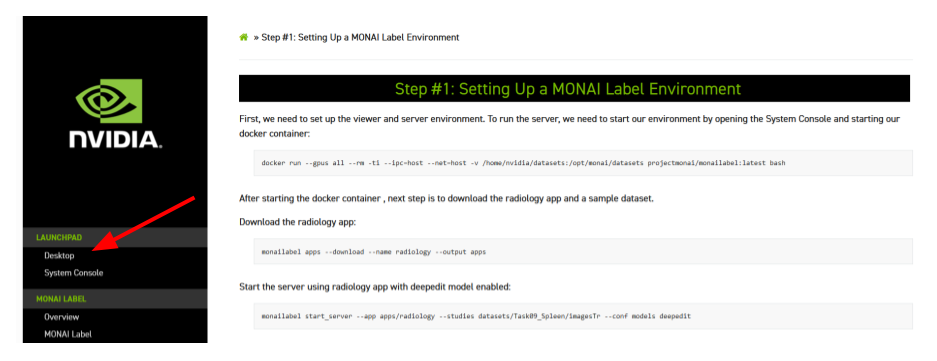
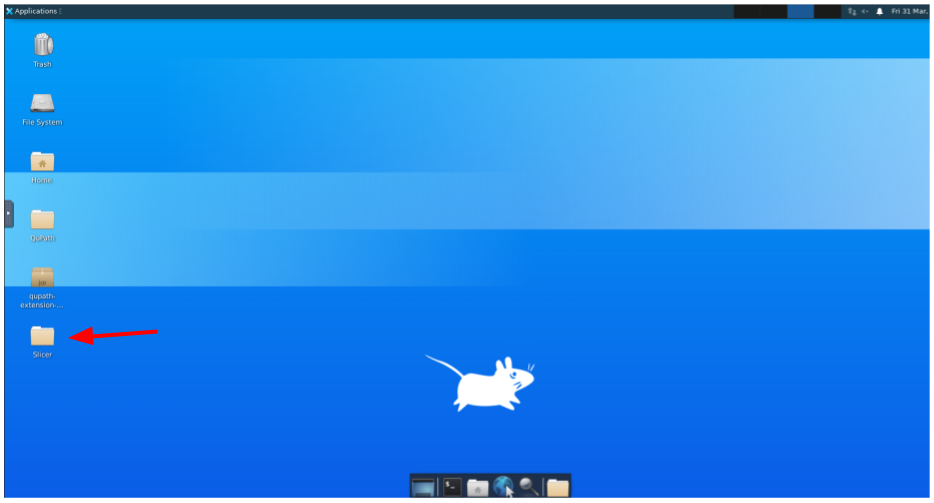
Once inside the Desktop, open the Slicer folder:
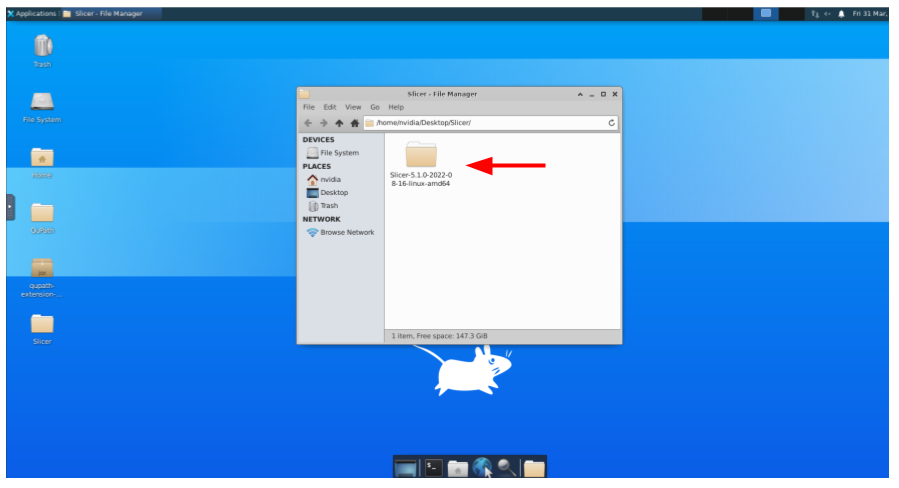
And then the subfolder:
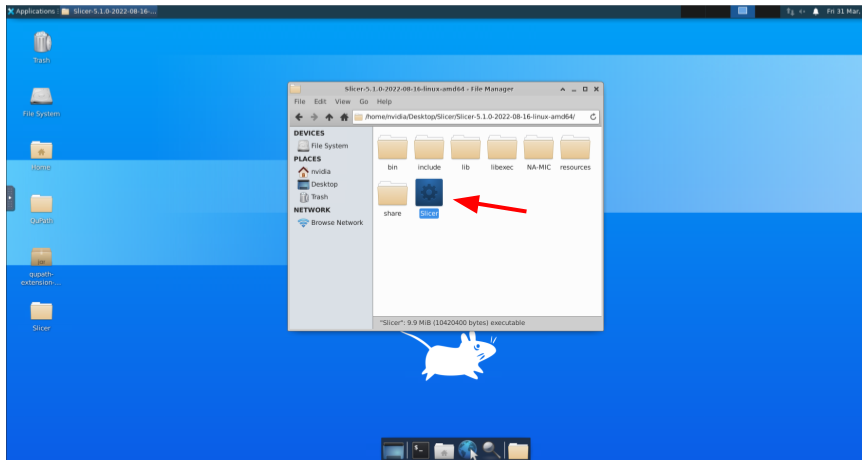
And double-click to the Slicer application:
Once Slicer is opened, go to View -> Extension Manager -> Install Extensions -> Active Learning.
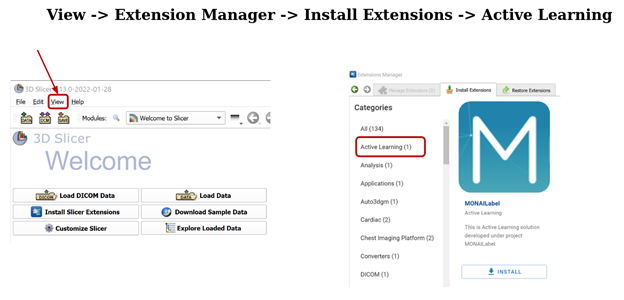
You have to relaunch 3D Slicer after installing the MONAI Label module and then access it through the Modules -> Active Learning menu.
Once the module is installed, type this IP address and port http://localhost:8000 into the MONAI Label Server box and click on the green circle icon, as it is shown in the following image:
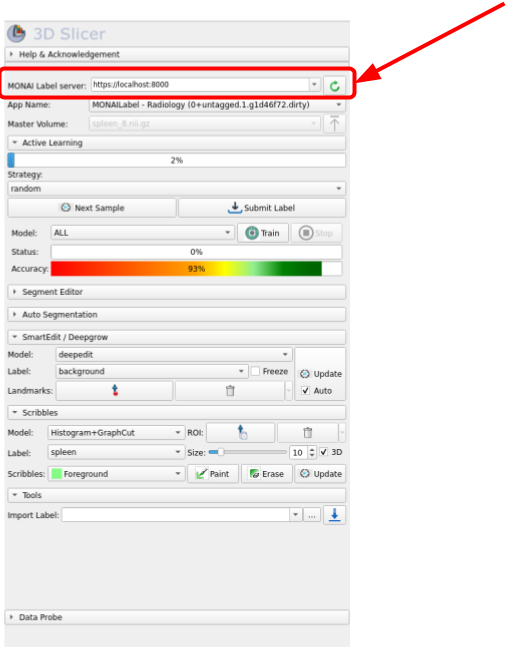
Once connected to the server, the next step is to fetch one of the unlabeled volumes. For this, click on Next Sample as shown in the image below:
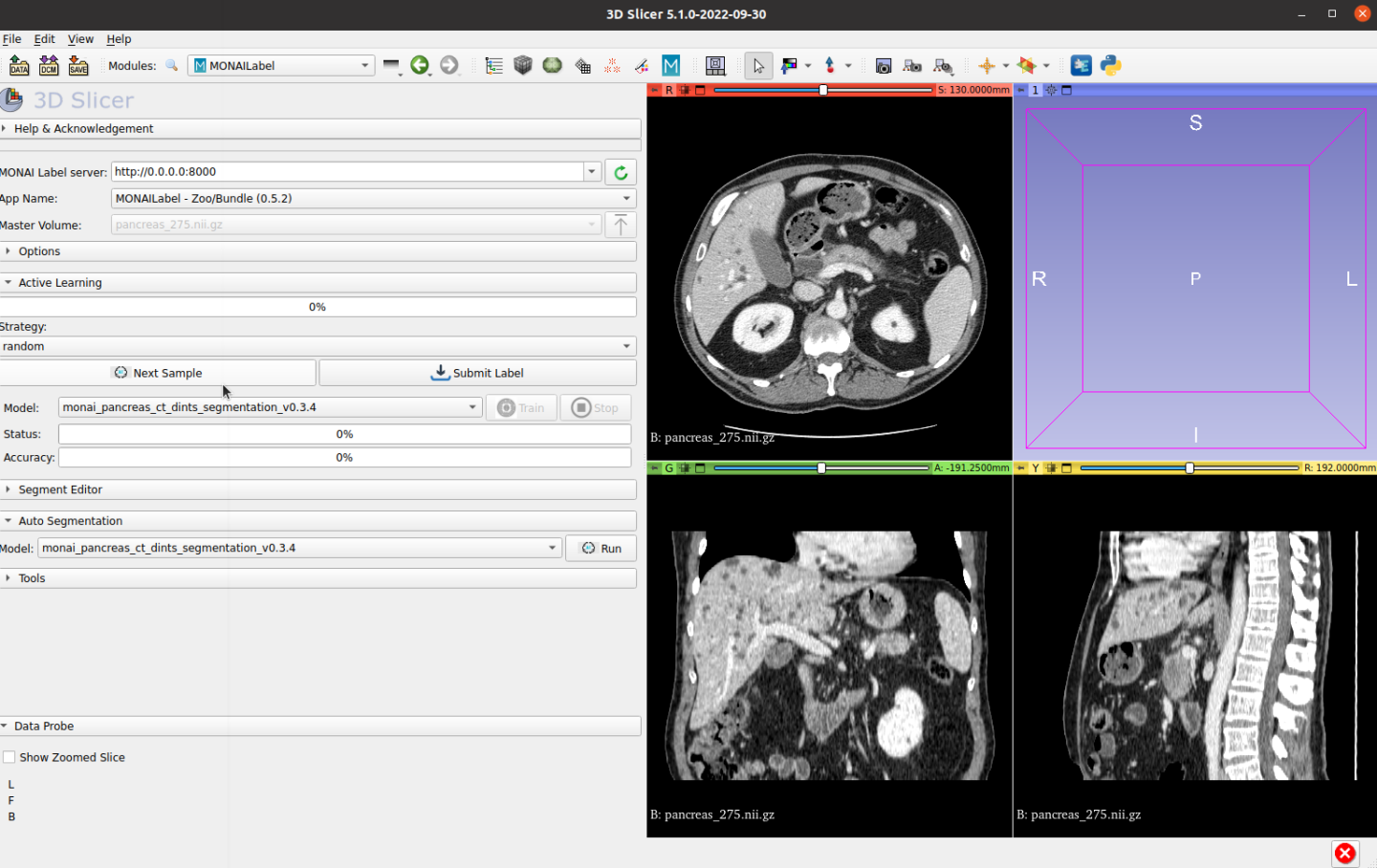
After fetching an unlabeled sample, users can run automatic inference to get both pancreas and tumor:
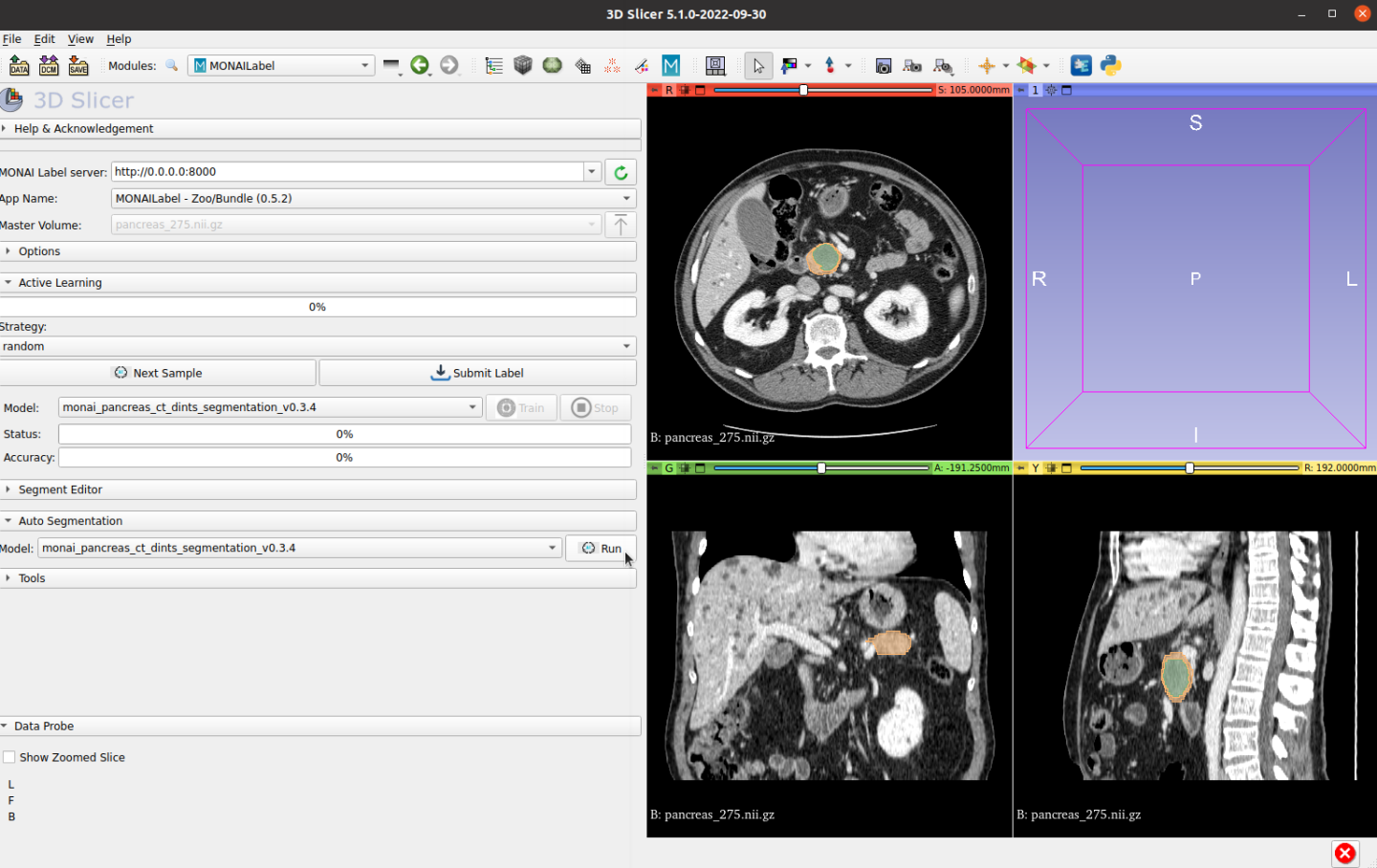
Then, edit the prediction if needed:
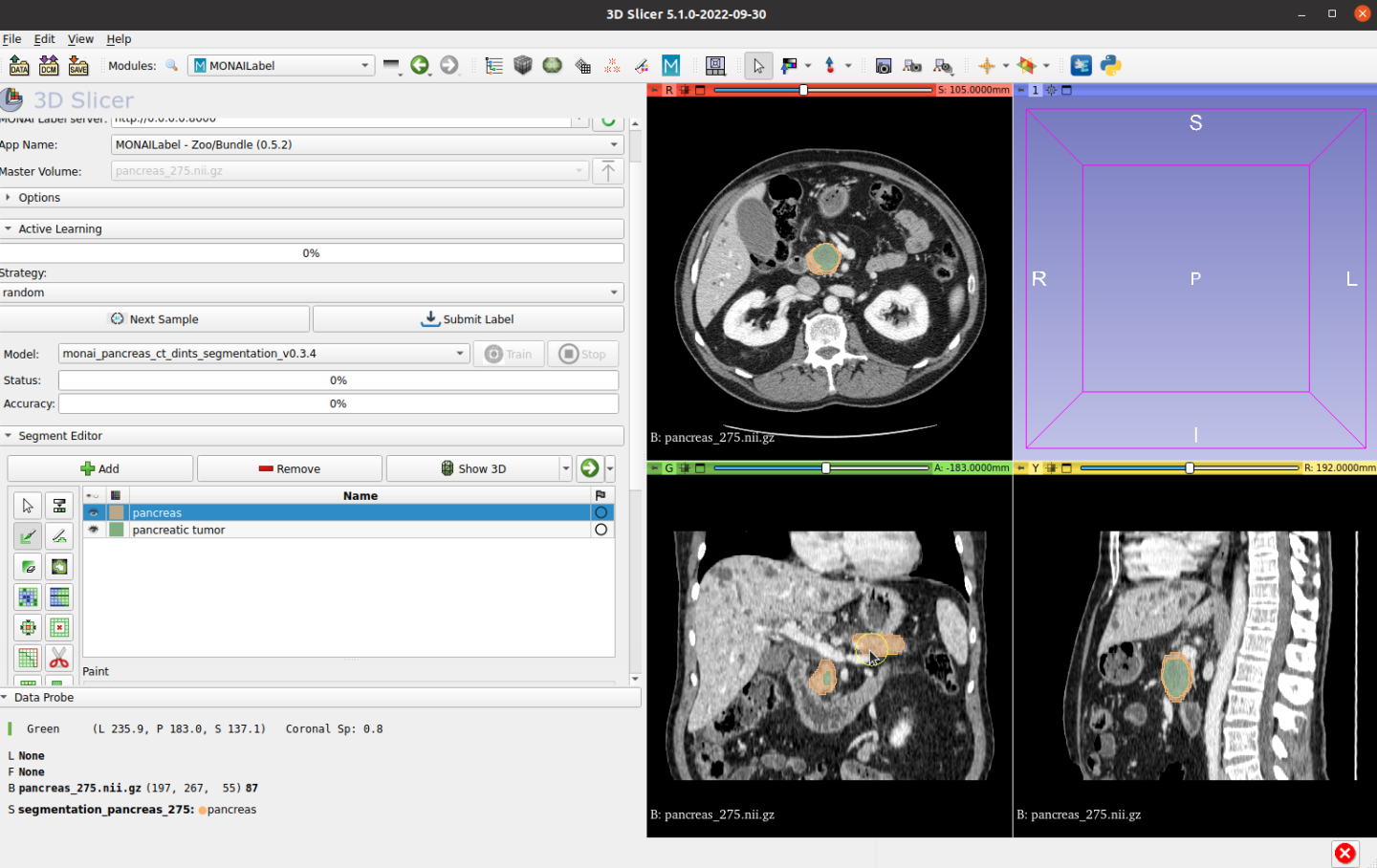
And click Submit Label to save the ground truth label to the file system:
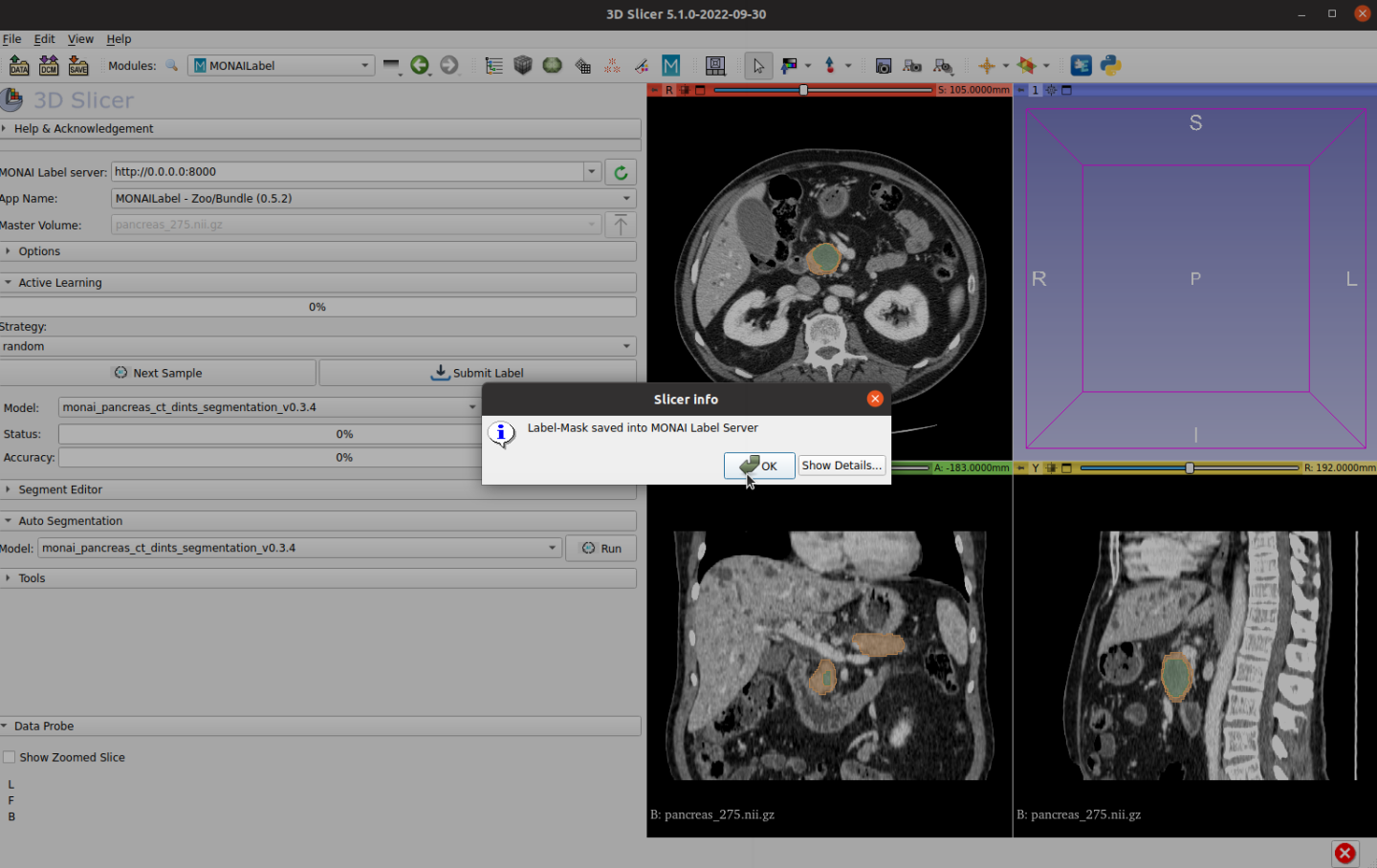
MONAI Label offers active learning strategies and can interactively fine-tune models. Users can train their models once new annotated labels are saved, with any number of finished labels. MONAI Label server set train and validation split percentage to 0.2 by default. It’s recommended to label five or more before running training. Click Train button, MONAI Label server will fetch the saved final ground truth label and fine-tune the prior model.
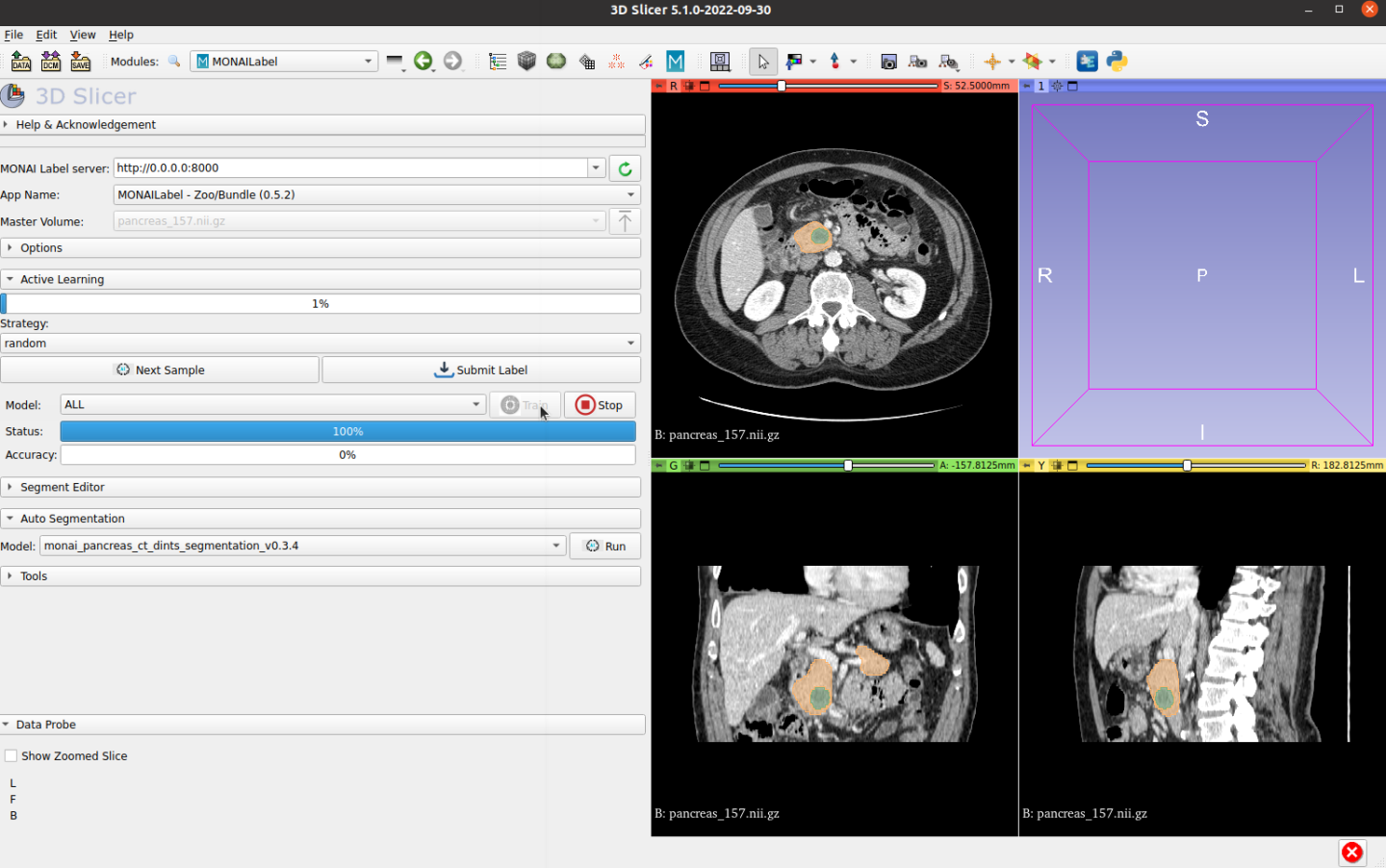
The active learning process will select unlabeled batch data, users can choose strategies in the Slicer UI.
In the second round of labeling, when the next image is selected, those trained images will be marked as labeled data, and won’t be selected. Active learning strategies such as “first/random” will be used for selecting which unlabeled data.
Iteratively label and train samples until all unlabeled data are annotated. The model will be fine-tuned with all labeled data at the end.
If you are done with the MONAI Label lab, just close the browser window/tab.