Step #1: Setting Up a MONAI Label Environment
The advanced feature of the endoscopic application is to actively select and fine-tune models.
If you’ve started the Docker container for previous apps (Radiology or Pathology), the first step for this use case is to stop the previous MONAI Label server by pressing Ctrl+C and then exit from the docker container by typing exit
First, we need to create a username and password for the CVAT and then set up the MONAI Label server environment.
If a superuser has been created for Use case 1 of the Endoscopy application, please skip this step.
If not, we need to start our environment by opening the System Console and running the following command to create a username and password:
cd /home/nvidia/cvat & docker exec -it cvat bash -ic 'python3 ~/manage.py createsuperuser'
The System Console can be found on the top left on this page:
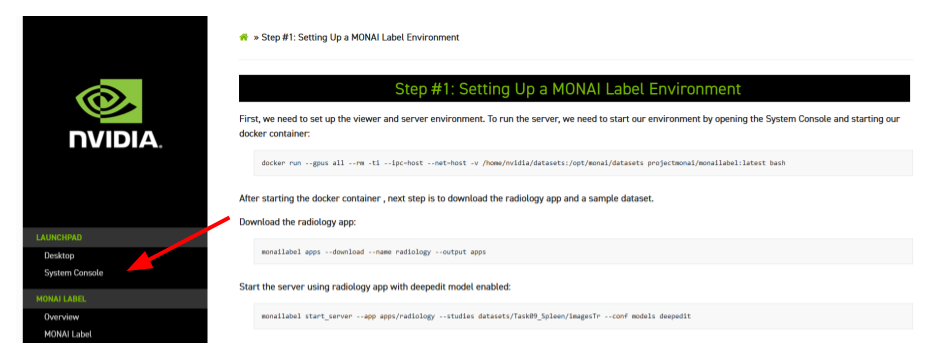
Once the username and password have been created, we need to start our docker container:
docker run --gpus all --rm -ti --ipc=host --net=host -v /home/nvidia/datasets:/opt/monai/datasets projectmonai/monailabel:latest bash
Skip this step of starting the docker container if you’ve already started the container for a previous app. There’s no need to start a new container if you already started one for another app. Stop the previous server by pressing Ctrl+C and go to the next step.
After the docker container starts, the next step is to download the pathology app and a sample dataset.
Download the endoscopy app:
monailabel apps --download --name endoscopy --output apps
Unzip the endoscopy sample dataset file:
unzip datasets/endoscopy_frames.zip -d datasets/endoscopy_frames
Start the server using the endoscopy app for active learning. Before that, please replace the text with the username and password previously created:
export MONAI_LABEL_DATASTORE=cvat
export MONAI_LABEL_DATASTORE_URL=http://127.0.0.1:8080
export MONAI_LABEL_DATASTORE_USERNAME=REPLACE_THIS_BY_YOUR_USERNAME
export MONAI_LABEL_DATASTORE_PASSWORD=REPLACE_THIS_BY_YOUR_PASSWORD
monailabel start_server \
--app apps/endoscopy \
--studies datasets/endoscopy_frames \
--conf models tooltracking \
--conf epistemic_enabled true \
--conf epistemic_top_k 10 \
--conf auto_finetune_models tooltracking \
--conf auto_finetune_check_interval 30
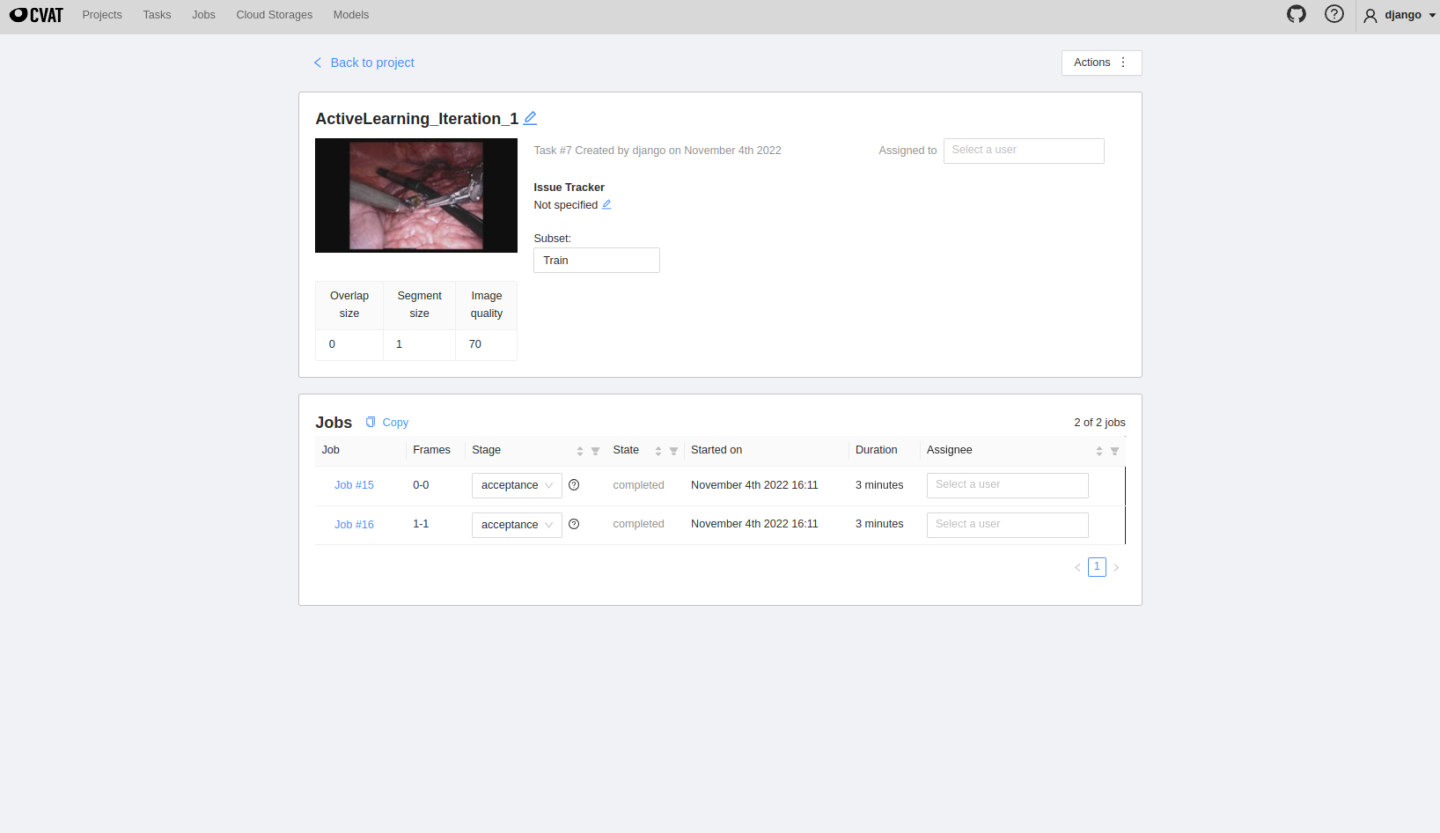
MONAI Label server will automatically upload selected images to CVAT. The automatically created project and tasks can be seen here:
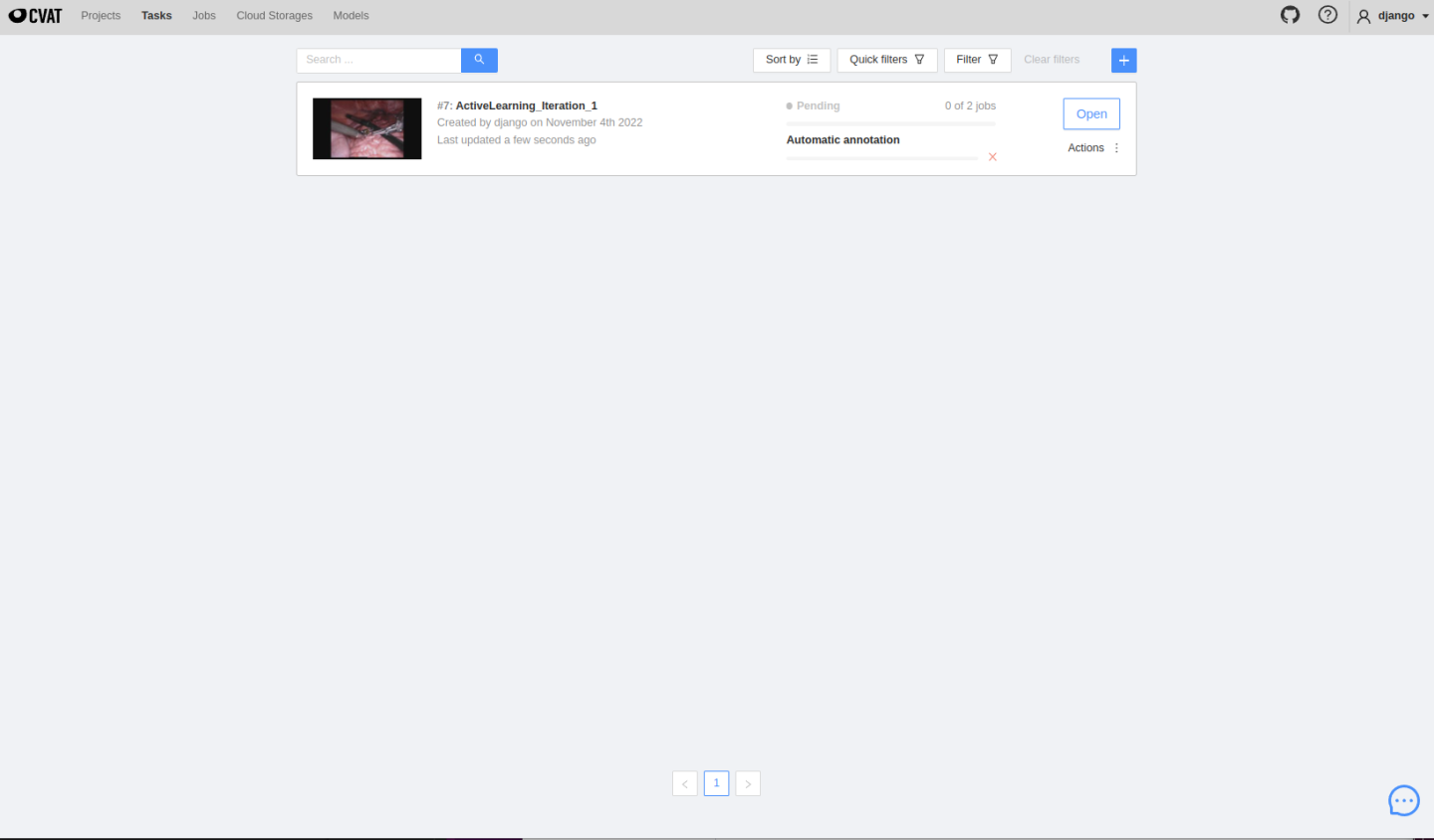
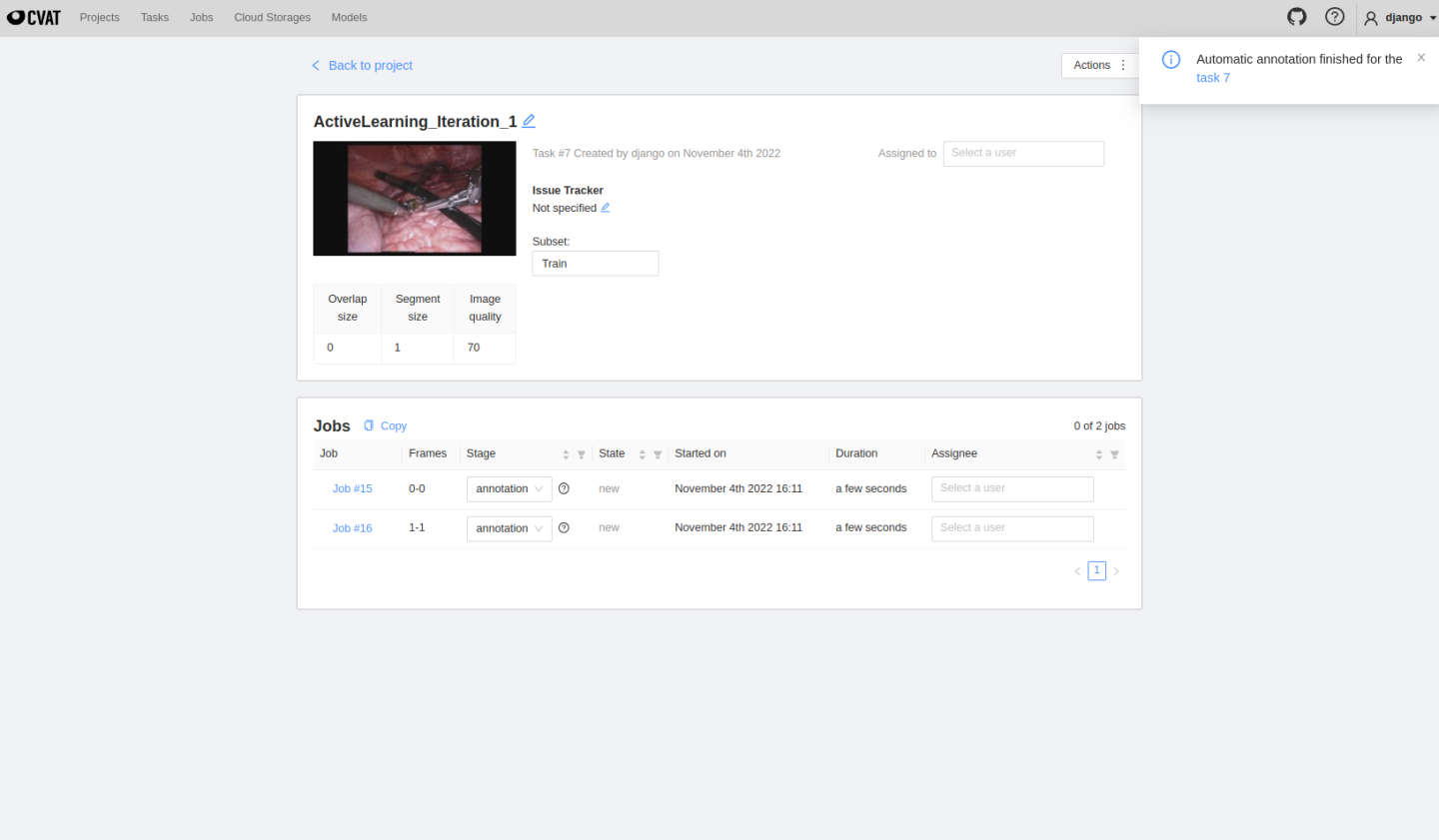
The created tasks will run inference automatically, and users can see predicted masks on the job page, edit them and follow the same annotating process presented before in Usecase 1. Once finished, click save, change the job state and finish the job.
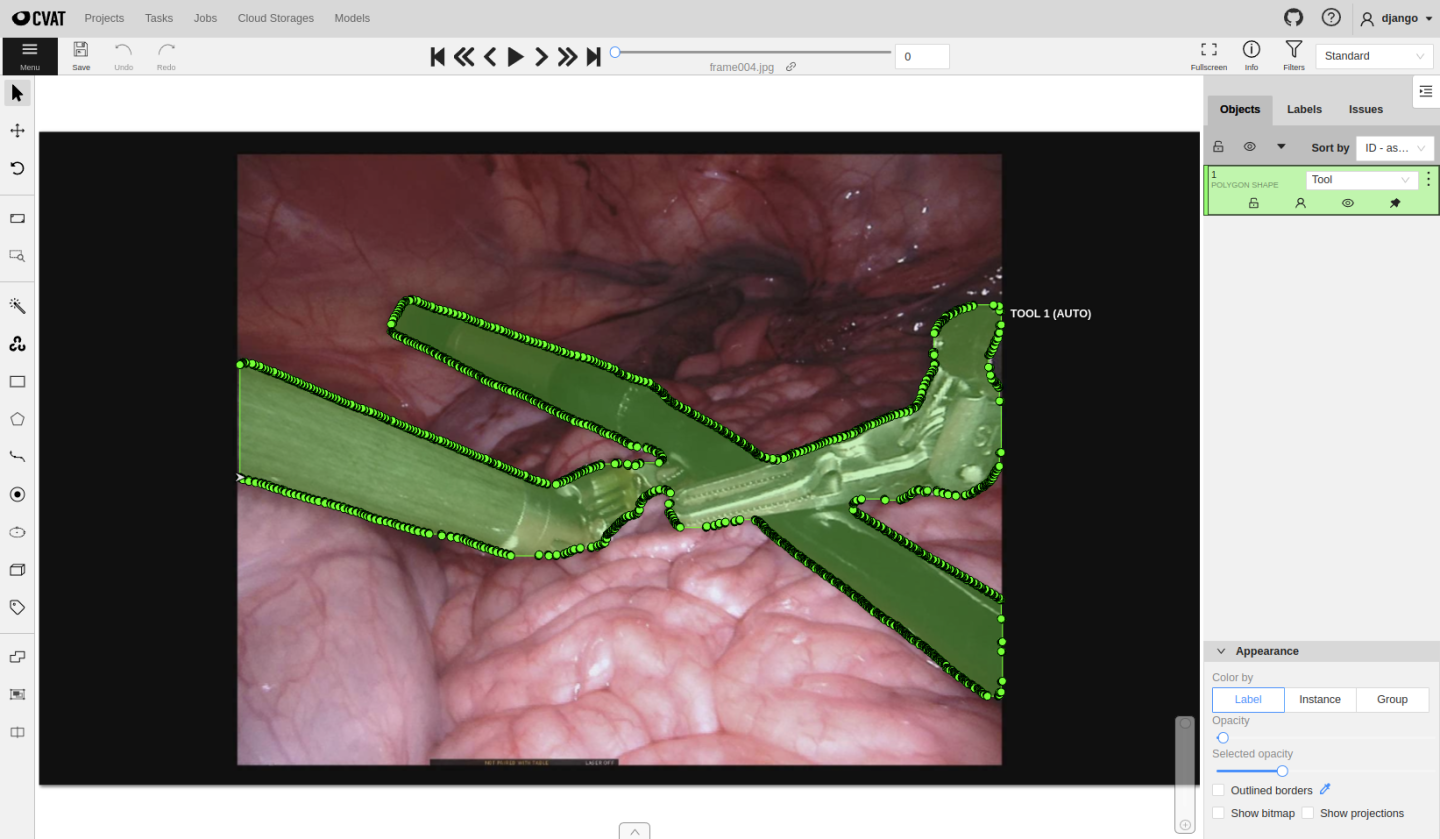
Continue to annotate until all created jobs are completed. MONAI Label will monitor the datastore status to check whether all jobs are in “completed” status.
Once all jobs are completed, the MONAI Label server will automatically trigger model fine-tuning and train the model with newly generated labels.
We have demonstrated how MONAI Label can be used for an advanced active learning workflow for endoscopic applications. Using CVAT and Nuclio deployment, the MONAI Label has the leading functionality of computer-assisted intervention tasks.
Now that you have a working MONAI Label implementation, work through our Quick Start Guide and/or the resources available on the Next Steps page.
If you are done with the MONAI Label lab, just close the browser window/tab.