Bring your own components (BYOC)
Clara allows researchers to solve new/different problems and innovate by writing their own components in a modular way. In order to do this, users can write their own components in python files then point to these files in the train_config.json file by providing the paths for the new components.
Below is a list of different components users can add. For examples, please see this Jupyter Notebook on Bring your own components (BYOC)
Data pipelines
A data pipeline contains a chain of transforms that are applied to the input image and label data to produce the data in the format required by the model.
Data pipelines produce batched data items during training. Typically, two data pipelines are used: one for producing training data, another producing validation data.
Model
The model component implements the neural network. It produces prediction for inputs.
Loss
The loss component implements a loss function, typically based on the prediction from the model and corresponding label data.
Optimizer
The optimizer component implements the training optimization algorithm for finding minimal loss during training.
Metrics
These components are used to dynamically measure the quality of the model during training on different aspects. Metric values are computed based on values of tensors. There are two kinds of metric components: training metrics, and validation metrics.
A training metric is a graph-building component that adds computational operations to the training graph, which produce tensors for metric computation.
Validation metrics implement algorithms to compute values for different aspects of the model, based on the values of tensors in the graph.
Structure of training graph
This diagram shows the overall structure of the training graph. It shows how the components are related. The blue ovals represent placeholders.
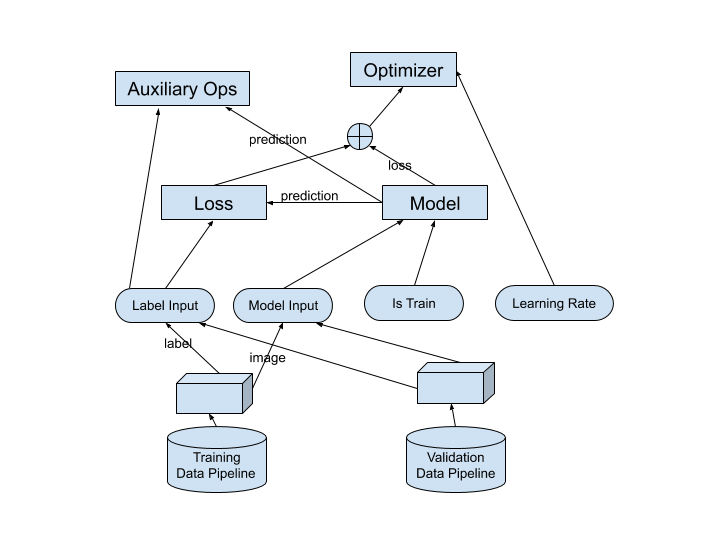
These components are built in this order:
Training Data Pipeline
Validation Data Pipeline
Placeholders
Model
Loss
Optimizer
Metrics
You can use the predefined models offered by NVIDIA, or you can choose to use your own model architecture when configuring a training workflow, provided your model follows our model development guidelines.
Model API specification
The model must conform to the API spec.
import tensorflow as tf
from ai4med.common.graph_component import GraphComponent
from ai4med.common.build_ctx import BuildContext
class Model(GraphComponent):
"""Base class of Models
Args:
None
Returns:
Prediction results
"""
def __init__(self):
GraphComponent.__init__(self)
def get_loss(self):
"""Get the additional loss function in AHNet model.
Args:
None
Returns:
Loss function
"""
return 0
def get_update_ops(self):
"""Get the update_ops for Batch Normalization.
The method "tf.control_dependencies" allow the operations used as inputs
of the context manager are run before the operations defined inside the
context manager. So we use "update_ops" to implement Batch Normalization.
Args:
None
Returns:
Update operations
"""
return tf.get_collection(tf.GraphKeys.UPDATE_OPS)
def get_predictions(self, inputs, is_training, build_ctx: BuildContext):
"""Forward computation process of model for both training and inference.
Args:
inputs (tf.Tensor): input data for the AHNet model
is_training (bool): in training process or not
build_ctx(BuildContext): reserved argument for future features
Returns:
Prediction results
"""
raise NotImplementedError('Class{}does not implement get_predictions'.format(
self.__class__.__name__))
def build(self, build_ctx: BuildContext):
"""Connect model with graph.
Args:
build_ctx: specified graph context
Returns:
Prediction results
"""
inputs = build_ctx.must_get(BuildContext.KEY_MODEL_INPUT)
is_training = build_ctx.must_get(BuildContext.KEY_IS_TRAIN)
return self.get_predictions(inputs, is_training, build_ctx)
Your model must extend the class Model and implement the required abstract methods.
get_predictions method
This method is required and is called during the construction of the computation graph. It must return a prediction tensor, as shown in the diagram above.
The inputs argument is the model input placeholder of the model.
The build_ctx argument is a dict that holds the data objects that are already built (see the component building order above). You can use them in the construction of your model. Specifically, by the time the get_predictions method is called, data pipelines and placeholders are already built, and the build_ctx contains the following objects:
data_property – properties about the input data such as data format (channels_first, channels_last), number of image channels, number of label channels, etc.
model_input – the placeholder for model input
label_input – the placeholder for label input
learning_rate – the placeholder for learning rate
is_train – the placeholder for is training flag
get_loss method
The get_loss method is called during the construction of the computation graph. You can override the default implementation of this method (which returns 0) if you want to return a model-specific loss. This loss is added to the result of the regular loss component.
get_update_ops method
You can also provide model-specific update ops using this method. The update ops will be used as the dependency for the Optimizer’s minimize operation.
Model creation
Clara manages components with a create and use strategy. Components are first configured and created based on the configuration parameters.
The configuration parameters are passed to the component’s construction method, __init__ , to get the component created. Since the parameters are defined at configuration time, they can only be simple static values (vs. dynamically created values such as tensors). Once the components are all created, workflow engine will start the graph construction process, which will invoke each component’s graph-building methods.
When creating your own model, you must follow this strategy: the __init__ method of the model class must only expect configuration parameters.
Examples
Extend the model class
To extend the model class, first, define your model as a subclass of the Model class:
import tensorflow as tf
from ai4med.components.models.model import Model
Create the model
The model’s constructor must only accept configurable parameters. Keep them in instance variables.
import tensorflow as tf
from ai4med.components.models.model import Model
class CustomNetwork(Model):
def __init__(self, num_classes,
factor=32,
training=False,
data_format='channels_first',
final_activation='linear'):
Model.__init__(self)
self.model = None
self.num_classes = num_classes
self.factor = factor
self.training = training
self.data_format = data_format
self.final_activation = final_activation
if data_format == 'channels_first':
self.channel_axis = 1
elif data_format == 'channels_last':
self.channel_axis = -1
def network(self, inputs, training, num_classes, factor, data_format, channel_axis):
# very shallow Unet Network
with tf.variable_scope('CustomNetwork'):
conv1_1 = tf.keras.layers.Conv3D(factor, 3, padding='same', data_format=data_format, activation='relu')(inputs)
conv1_2 = tf.keras.layers.Conv3D(factor * 2, 3, padding='same', data_format=data_format, activation='relu')(conv1_1)
pool1 = tf.keras.layers.MaxPool3D(pool_size=(2, 2, 2), strides=2, data_format=data_format)(conv1_2)
conv2_1 = tf.keras.layers.Conv3D(factor * 2, 3, padding='same', data_format=data_format, activation='relu')(pool1)
conv2_2 = tf.keras.layers.Conv3D(factor * 4, 3, padding='same', data_format=data_format, activation='relu')(conv2_1)
unpool1 = tf.keras.layers.UpSampling3D(size=(2, 2, 2), data_format=data_format)(conv2_2)
unpool1 = tf.keras.layers.Concatenate(axis=channel_axis)([unpool1, conv1_2])
conv7_1 = tf.keras.layers.Conv3D(factor * 2, 3, padding='same', data_format=data_format, activation='relu')(unpool1)
conv7_2 = tf.keras.layers.Conv3D(factor * 2, 3, padding='same', data_format=data_format, activation='relu')(conv7_1)
output = tf.keras.layers.Conv3D(num_classes, 1, padding='same', data_format=data_format)(conv7_2)
if str.lower(self.final_activation) == 'softmax':
output = tf.nn.softmax(output, axis=channel_axis, name='softmax')
elif str.lower(self.final_activation) == 'sigmoid':
output = tf.nn.sigmoid(output, name='sigmoid')
elif str.lower(self.final_activation) == 'linear':
pass
else:
raise ValueError(
'Unsupported final_activation, it must of one (softmax, sigmoid or linear), but provided:' + self.final_activation)
return output
# additional custom loss
def loss(self):
return 0
def get_predictions(self, inputs, training, build_ctx=None):
self.model = self.network(
inputs=inputs,
training=training,
num_classes=self.num_classes,
factor=self.factor,
data_format=self.data_format,
channel_axis=-1
)
return self.model
def get_loss(self):
return self.loss()
Implement methods
Define the get_predictions method.
Optional methods
Optionally, you can define the get_loss method and the get_update_ops method for the model.
Configuration
Once your model is developed following the guidelines, you can use it in the training workflow with the following steps:
Locate the section for model in the training config JSON file.
Specify the path to your model’s class.
Specify all required init parameters in the args section.
Make sure that the specified model class path is in PYTHONPATH.
Here is sample training config file:
{
"epochs": 1240,
"num_training_epoch_per_valid": 20,
"learning_rate": 1e-4,
"multi_gpu": false,
"train":
{
"loss":
{
"name": "Dice"
},
"optimizer":
{
"name": "Adam"
},
...
"model":
{
"path": "pythonPathToYourModelClass",
"args": {
"num_classes": 2,
"factor": 8,
"final_activation": "softmax"
}
},
...
}
...
}
The pythonPathToYourModelClass must be accessible through PYTHONPATH.
For example, if pythonPathToYourModelClass is defined as: foo.bar.FancyNet and the class FancyNet is implemented in
/project/deeplearn/foo/bar.py
then, PYTHONPATH must include
/project/deeplearn
Users can write their own transformations for data augmentation. Transform components are now designed to utilize a
TransformContext that stores MedicalImages
as fields to keep ShapeFormat information with the image data. See MedicalImages with ShapeFormat for details.
Below is an example to add/subtract a random constant to the image as an augmentation transformation. A transform
template can be downloaded here: transforms_template.py.
import numpy as np
# note the ai4med here
from ai4med.common.medical_image import MedicalImage
from ai4med.common.transform_ctx import TransformContext
from ai4med.components.transforms.multi_field_transformer import MultiFieldTransformer
class MyAddRandomConstant(MultiFieldTransformer):
def __init__(self, fields, magnitude, dtype=np.float32):
# fields specifies the names of the image fields in the data dict that you want to add constant to
MultiFieldTransformer.__init__(self, fields)
self.dtype = dtype
self.magnitude = magnitude
def transform(self, transform_ctx):
for field in self.fields:
offset = (np.random.rand() * 2.0 - 1.0) * self.magnitude
# get the MedicalImage using field
img = transform_ctx.get_image(field)
# get_data give us a numpy array of data
result = img.get_data() + offset
# create a new MedicalImage use new_image() method
# which will carry over the properties of the original image
result_img = img.new_image(result, img.get_shape_format())
# set the image back in transform_ctx
transform_ctx.set_image(field, result_img)
return transform_ctx
def is_deterministic(self):
""" This is not a deterministic transform.
Returns:
False (bool)
"""
return False
By default, is_deterministic returns true, so it is important to override this if the transform is not deterministic.
Then the user would add the following lines in train_config.json:
"pre_transforms":
[
{
"name": "MyAddRandomConstant",
"path": "CustomCode.MyAddRandomConstant",
"args": {
"fields": ["image", "label"],
"magnitude": 5
}
},
]
A Data Loader is a Transformation, but it is typically at the beginning of the chain of transforms. Also, the contents of the input data are not MedicalImages but usually the file paths to the data. Users can write their own data loaders to read different types of input such as png, Jpeg, matlab files, raw images, etc. They can process it as they desire and extract additional information such as affine or spacing to store as properties of a MedicalImage before finally placing the MedicalImage as a field in the TransformContext.
Below is an example for loading numpy files. Note that ShapeFormat is used as explained in MedicalImages with ShapeFormat.
import numpy as np
import logging
from ai4med.common.constants import ImageProperty
from ai4med.common.medical_image import MedicalImage
from ai4med.common.shape_format import ShapeFormat
from ai4med.common.transform_ctx import TransformContext
from ai4med.utils.dtype_utils import str_to_dtype
from ai4med.components.transforms.multi_field_transformer import MultiFieldTransformer
class MyNumpyReader(object):
"""Reads Numpy files.
Args:
dtype: Type for data to be loaded.
"""
def __init__(self, dtype=np.float32):
self._logger = logging.getLogger(self.__class__.__name__)
self._dtype = dtype
def read(self, file_name, shape: ShapeFormat):
assert shape, "Please provide a valid shape."
assert file_name, "Please provide a filename."
if isinstance(file_name, (bytes, bytearray)):
file_name = file_name.decode('UTF-8')
data = np.load(file_name, allow_pickle=True).astype(self._dtype)
assert len(data.shape) == shape.get_number_of_dims(), \
"Dims of loaded data and provided shape don't match."
img = MedicalImage(data, shape)
img.set_property(ImageProperty.ORIGINAL_SHAPE, data.shape)
img.set_property(ImageProperty.FILENAME, file_name)
return img
class MyNumpyLoader(MultiFieldTransformer):
"""Load Image from Numpy files.
Args:
shape (ShapeFormat): Shape of output image.
dtype : Type for output data.
"""
def __init__(self, fields, shape, dtype="float32"):
MultiFieldTransformer.__init__(self, fields=fields)
self._dtype = str_to_dtype(dtype)
self._shape = ShapeFormat(shape)
self._reader = MyNumpyReader(self._dtype)
def transform(self, transform_ctx: TransformContext):
for field in self.fields:
file_name = transform_ctx[field]
transform_ctx.set_image(field, self._reader.read(file_name, self._shape))
return transform_ctx
Update the config_train.json file:
{
"name": "MyNumpyLoader",
"path": "CustomCode.MyNumpyLoader",
"args": {
"fields": [
"image",
"label"
],
"shape": "DHW"
}
},
Because np.load returns a N-dimension array, and it is impossible to know if it is in “CHW”, “DHW” or any other format, users will have to provide “shape” argument in order for loader to output correct shape format.
Note that if the N-dimension array from np.load is not in a standard shape formats, as listed below, users need to implement some operations to covert the N-dimension array into one of the standard shape formats.
# 3D
DHW = 'DHW'
DHWC = 'DHWC'
CDHW = 'CDHW'
NDHW = 'NDHW'
NDHWC = 'NDHWC'
NCDHW = 'NCDHW'
# 2D
HW = 'HW'
HWC = 'HWC'
CHW = 'CHW'
NHW = 'NHW'
NHWC = 'NHWC'
NCHW = 'NCHW'
User write the loss in a python file by implementing the Loss class as follows.
import tensorflow as tf
from ai4med.components.losses.loss import Loss
def dice_loss(predictions,
targets,
data_format='channels_first',
skip_background=False,
squared_pred=False,
jaccard=False,
smooth=1e-5,
top_smooth=0.0,
is_onehot_targets=False):
"""Compute average Dice loss between two tensors.
5D tensors (for 3D images) or 4D tensors (for 2D images).
Args:
predictions (Tensor): Tensor of Predicted segmentation output (e.g NxCxHxWxD)
targets (Tensor): Tensor of True segmentation values. Usually has 1 channel dimension (e.g. Nx1xHxWxD),
where each element is an index indicating class label.
Alternatively it can be a one-hot-encoded tensor of the shape NxCxHxWxD,
where each channel is binary (or float in interval 0..1) indicating
the probability of the corresponding class label
data_format (str): channels_first (default) or channels_last
skip_background (bool): skip dice computation on the first channel of the predicted output or not
squared_pred (bool): use squared versions of targets and predictions in the denominator or not
jaccard (bool): compute Jaccard Index (soft IoU) instead of dice or not
smooth (float): denominator constant to avoid zero division (default 1e-5)
top_smooth (float): experimental, nominator constant to avoid zero final loss when targets are all zeros
is_onehot_targets (bool): the targets are already One-Hot-encoded or not
Returns:
tensor of one minus average dice loss
"""
is_channels_first = (data_format == 'channels_first')
ch_axis = 1 if is_channels_first else -1
n_channels_pred = predictions.get_shape()[ch_axis].value
n_channels_targ = targets.get_shape()[ch_axis].value
n_len = len(predictions.get_shape())
print('dice_loss targets', targets.get_shape().as_list(),
'predictions', predictions.get_shape().as_list(),
'targets.dtype', targets.dtype,
'predictions.dtype', predictions.dtype)
print('dice_loss is_channels_first:', is_channels_first,
'skip_background:', skip_background,
'is_onehot_targets', is_onehot_targets)
# Sanity checks
if skip_background and n_channels_pred == 1:
raise ValueError("There is only 1 single channel in the predicted output, and skip_zero is True")
if skip_background and n_channels_targ == 1 and is_onehot_targets:
raise ValueError("There is only 1 single channel in the true output (and it is is_onehot_true), "
"and skip_zero is True")
if is_onehot_targets and n_channels_targ != n_channels_pred:
raise ValueError("Number of channels in target{}and pred outputs{}"
"must be equal to use is_onehot_true == True".format(
n_channels_targ, n_channels_pred))
# End sanity checks
if not is_onehot_targets:
# if not one-hot representation already
# remove singleton (channel) dimension for true labels
targets = tf.cast(tf.squeeze(targets, axis=ch_axis), tf.int32)
targets = tf.one_hot(targets, depth=n_channels_pred, axis=ch_axis,
dtype=tf.float32, name="loss_dice_targets_onehot")
if skip_background:
# if skipping background, removing first channel
targets = targets[:, 1:] if is_channels_first else targets[..., 1:]
predictions = predictions[:, 1:] if is_channels_first else predictions[..., 1:]
# reducing only spatial dimensions (not batch nor channels)
reduce_axis = list(range(2, n_len)) if is_channels_first else list(range(1, n_len - 1))
intersection = tf.reduce_sum(targets * predictions, axis=reduce_axis)
if squared_pred:
# technically we don't need this square for binary true values
# (but in cases where true is probability/float, we still need to square
targets = tf.square(targets)
predictions = tf.square(predictions)
y_true_o = tf.reduce_sum(targets, axis=reduce_axis)
y_pred_o = tf.reduce_sum(predictions, axis=reduce_axis)
denominator = y_true_o + y_pred_o
if jaccard:
denominator -= intersection
f = (2.0 * intersection + top_smooth) / (denominator + smooth)
# # If only compute dice for present label, mask out data-label that are not present
# if only_present:
# dice_mask = tf.logical_not(tf.equal(label_sum, 0))
# dice = tf.boolean_mask(dice, dice_mask)
f = tf.reduce_mean(f) # final reduce_mean across batches and channels
return 1 - f
class MyClonedDiceLoss(Loss):
"""Compute average Dice loss between two tensors.
5D tensors (for 3D images) or 4D tensors (for 2D images).
Args:
data_format (str): channels_first (default) or channels_last
skip_background (bool): skip dice computation on the first channel of the predicted output or not
squared_pred (bool): use squared versions of targets and predictions in the denominator or not
jaccard (bool): compute Jaccard Index (soft IoU) instead of dice or not
smooth (float): denominator constant to avoid zero division (default 1e-5)
top_smooth (float): experimental, nominator constant to avoid zero final loss when targets are all zeros
is_onehot_targets (bool): the targets are already One-Hot-encoded or not
Returns:
tensor of one minus average dice loss
"""
def __init__(self,
data_format='channels_first',
skip_background=False,
squared_pred=False,
jaccard=False,
smooth=1e-5,
top_smooth=0.0,
is_onehot_targets=False):
Loss.__init__(self)
self.data_format = data_format
self.skip_background = skip_background
self.squared_pred = squared_pred
self.jaccard = jaccard
self.smooth = smooth
self.top_smooth = top_smooth
self.is_onehot_targets = is_onehot_targets
def get_loss(self, predictions, targets, build_ctx=None):
"""Compute dice loss for tf variable
Args:
predictions (Tensor): outputs of the network
targets (Tensor): target integer labels
build_ctx: specified graph context
Returns:
tensor of dice loss
"""
return dice_loss(predictions, targets,
data_format=self.data_format,
skip_background=self.skip_background,
squared_pred=self.squared_pred,
jaccard=self.jaccard,
smooth=self.smooth,
top_smooth=self.top_smooth,
is_onehot_targets=self.is_onehot_targets)
When additional properties about the data are required in order to compute the loss, they can be retrieved via build_ctx. The following example retrieves the KEY_DATA_PROP from build_ctx.
data_prop = build_ctx.must_get(BuildContext.KEY_DATA_PROP)
Then users can add the loss in the training section of the train_config.json file:
"train":
{
"loss": {
"name": "MyClonedDiceLoss",
"path": "CustomCode.MyClonedDiceLoss",
"args": {
"skip_background": false
}
},
import numpy as np
from ai4med.components.metric import Metric
from ai4med.libs.metrics.metric_list import MetricList
class SampleMetricAverage(MetricList):
"""
Generic class for tracking averages of metrics. Expects that the elements in self._list
are scalar values that will be averaged
"""
def __init__(self, name, invalid_value=float('nan'), report_path=None):
MetricList.__init__(self, name,
invalid_value=invalid_value,
report_path=report_path)
def get(self):
if self._list is None or self._list.size == 0:
return 0
return np.mean(self._list)
class SampleComputeAverage(Metric):
def __init__(self, name, field,
invalid_value=float('nan'),
report_path=None,
do_summary=True,
do_print=True,
is_key_metric=False):
m = SampleMetricAverage(name, invalid_value, report_path)
Metric.__init__(self, m,
field=field,
do_summary=do_summary,
do_print=do_print,
is_key_metric=is_key_metric)
Update the train_config.json file:
"metrics": [
{
"name": "SampleComputeAverage",
"path": "CustomCode.SampleComputeAverage",
"args": {
"name": "metric_mean_value_1",
"field": "accuracy"
}
]
In this example, the metric subclasses Metric and instantiates SampleMetricAverage class, which can handle list of metrics.
The SampleMetricAverage must implement the get() method to return the computed metrics.