Coupled Spring Mass ODE System#
Introduction#
In this tutorial PhysicsNeMo Sym is used to solve a system of coupled ordinary differential equations. Since the APIs used for this problem have already been covered in a previous tutorial, only the problem description is discussed without going into the details of the code.
Note
This tutorial assumes that you have completed tutorial Introductory Example and have familiarized yourself with the basics of the PhysicsNeMo Sym APIs. Also, refer to tutorial 1D Wave Equation for information on defining new differential equations, and solving time dependent problems in PhysicsNeMo Sym.
Problem Description#
In this tutorial, a simple spring mass system as shown in Fig. 114 is solved. The systems shows three masses attached to each other by four springs. The springs slide along a frictionless horizontal surface. The masses are assumed to be point masses and the springs are massless. The problem is solved so that the masses (\(m's\)) and the spring constants (\(k's\)) are constants, but they can later be parameterized if you intend to solve the parameterized problem (Tutorial Parameterized 3D Heat Sink).
The model’s equations are given as below:
Where, \(x_1(t), x_2(t), \text{and } x_3(t)\) denote the mass positions along the horizontal surface measured from their equilibrium position, plus right and minus left. As shown in the figure, first and the last spring are fixed to the walls.
For this tutorial, assume the following conditions:
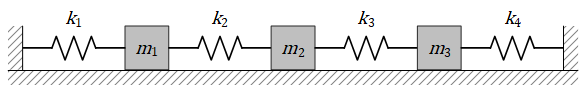
Fig. 114 Three masses connected by four springs on a frictionless surface#
Case Setup#
The case setup for this problem is very similar to the setup
for the tutorial 1D Wave Equation. You can define the
differential equations in spring_mass_ode.py and then define the
domain and the solver in spring_mass_solver.py.
Note
The python scripts for this problem can be found at examples/ode_spring_mass/.
Defining the Equations#
The equations of the system (21) can be coded using the sympy notation similar to tutorial 1D Wave Equation.
# SPDX-FileCopyrightText: Copyright (c) 2023 - 2024 NVIDIA CORPORATION & AFFILIATES.
# SPDX-FileCopyrightText: All rights reserved.
# SPDX-License-Identifier: Apache-2.0
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from sympy import Symbol, Function, Number
from physicsnemo.sym.eq.pde import PDE
class SpringMass(PDE):
name = "SpringMass"
def __init__(self, k=(2, 1, 1, 2), m=(1, 1, 1)):
self.k = k
self.m = m
k1 = k[0]
k2 = k[1]
k3 = k[2]
k4 = k[3]
m1 = m[0]
m2 = m[1]
m3 = m[2]
t = Symbol("t")
input_variables = {"t": t}
x1 = Function("x1")(*input_variables)
x2 = Function("x2")(*input_variables)
x3 = Function("x3")(*input_variables)
if type(k1) is str:
k1 = Function(k1)(*input_variables)
elif type(k1) in [float, int]:
k1 = Number(k1)
if type(k2) is str:
k2 = Function(k2)(*input_variables)
elif type(k2) in [float, int]:
k2 = Number(k2)
if type(k3) is str:
k3 = Function(k3)(*input_variables)
elif type(k3) in [float, int]:
k3 = Number(k3)
if type(k4) is str:
k4 = Function(k4)(*input_variables)
elif type(k4) in [float, int]:
k4 = Number(k4)
if type(m1) is str:
m1 = Function(m1)(*input_variables)
elif type(m1) in [float, int]:
m1 = Number(m1)
if type(m2) is str:
m2 = Function(m2)(*input_variables)
elif type(m2) in [float, int]:
m2 = Number(m2)
if type(m3) is str:
m3 = Function(m3)(*input_variables)
elif type(m3) in [float, int]:
m3 = Number(m3)
self.equations = {}
self.equations["ode_x1"] = m1 * (x1.diff(t)).diff(t) + k1 * x1 - k2 * (x2 - x1)
self.equations["ode_x2"] = (
m2 * (x2.diff(t)).diff(t) + k2 * (x2 - x1) - k3 * (x3 - x2)
)
self.equations["ode_x3"] = m3 * (x3.diff(t)).diff(t) + k3 * (x3 - x2) + k4 * x3
Here, each parameter \((k's \text{ and } m's)\) is written as a function and is substituted as a number if it’s constant. This will allow you to parameterize any of this constants by passing them as a string.
Solving the ODEs: Creating Geometry, defining ICs and making the Neural Network Solver#
Once you have the ODEs defined, you can easily form the constraints needed for optimization as seen in earlier tutorials.
This example, uses Point1D
geometry to create the point mass. You also have to define the time range of
the solution and create symbol for time (\(t\)) to define the
initial condition, etc. in the train domain. This code shows the
geometry definition for this problem. Note that this tutorial does not use the x-coordinate
(\(x\)) information of the point, it is only used to sample a
point in space. The point is assigned different values for variable
(\(t\)) only (initial conditions and ODEs over the time range). The code to
generate the nodes and relevant constraints is below:
# limitations under the License.
import numpy as np
from sympy import Symbol
import physicsnemo.sym
from physicsnemo.sym.hydra import instantiate_arch, PhysicsNeMoConfig
from physicsnemo.sym.solver import Solver
from physicsnemo.sym.domain import Domain
from physicsnemo.sym.geometry.primitives_1d import Point1D
from physicsnemo.sym.domain.constraint import (
PointwiseBoundaryConstraint,
)
from physicsnemo.sym.domain.validator import PointwiseValidator
from physicsnemo.sym.key import Key
from spring_mass_ode import SpringMass
@physicsnemo.sym.main(config_path="conf", config_name="config")
def run(cfg: PhysicsNeMoConfig) -> None:
# make list of nodes to unroll graph on
sm = SpringMass(k=(2, 1, 1, 2), m=(1, 1, 1))
sm_net = instantiate_arch(
input_keys=[Key("t")],
output_keys=[Key("x1"), Key("x2"), Key("x3")],
cfg=cfg.arch.fully_connected,
)
nodes = sm.make_nodes() + [sm_net.make_node(name="spring_mass_network")]
# add constraints to solver
# make geometry
geo = Point1D(0)
t_max = 10.0
t_symbol = Symbol("t")
Symbol("x")
time_range = {t_symbol: (0, t_max)}
# make domain
domain = Domain()
# initial conditions
IC = PointwiseBoundaryConstraint(
nodes=nodes,
geometry=geo,
outvar={"x1": 1.0, "x2": 0, "x3": 0, "x1__t": 0, "x2__t": 0, "x3__t": 0},
batch_size=cfg.batch_size.IC,
lambda_weighting={
"x1": 1.0,
"x2": 1.0,
"x3": 1.0,
"x1__t": 1.0,
"x2__t": 1.0,
"x3__t": 1.0,
},
parameterization={t_symbol: 0},
)
domain.add_constraint(IC, name="IC")
# solve over given time period
interior = PointwiseBoundaryConstraint(
nodes=nodes,
geometry=geo,
outvar={"ode_x1": 0.0, "ode_x2": 0.0, "ode_x3": 0.0},
batch_size=cfg.batch_size.interior,
parameterization=time_range,
)
domain.add_constraint(interior, "interior")
Next, you can define the validation data for this problem. The solution of this problem can be obtained analytically and the expression can be coded into dictionaries of numpy arrays for \(x_1, x_2, \text{and } x_3\). This part of the code is similar to the tutorial 1D Wave Equation.
# add validation data
deltaT = 0.001
t = np.arange(0, t_max, deltaT)
t = np.expand_dims(t, axis=-1)
invar_numpy = {"t": t}
outvar_numpy = {
"x1": (1 / 6) * np.cos(t)
+ (1 / 2) * np.cos(np.sqrt(3) * t)
+ (1 / 3) * np.cos(2 * t),
"x2": (2 / 6) * np.cos(t)
+ (0 / 2) * np.cos(np.sqrt(3) * t)
- (1 / 3) * np.cos(2 * t),
"x3": (1 / 6) * np.cos(t)
- (1 / 2) * np.cos(np.sqrt(3) * t)
+ (1 / 3) * np.cos(2 * t),
}
validator = PointwiseValidator(
nodes=nodes, invar=invar_numpy, true_outvar=outvar_numpy, batch_size=1024
)
domain.add_validator(validator)
Now that you have the definitions for the various constraints and domains complete, you can form the solver and run the problem. The code to do the same can be found below:
# make solver
slv = Solver(cfg, domain)
# start solver
slv.solve()
Once the python file is setup, you can solve the problem by executing the
solver script spring_mass_solver.py as seen in other tutorials.
Results and Post-processing#
The results for the PhysicsNeMo Sym simulation are compared against the
analytical validation data. You can see that the solution converges to
the analytical result very quickly. The plots can be created
using the .npz files that are created in the validator/ directory
in the network checkpoint.
Fig. 115 Comparison of PhysicsNeMo Sym results with an analytical solution.#