Deep Operator Network
This tutorial illustrates how to learn abstract operators using data-informed and physics-informed Deep operator network (DeepONet) in Modulus. In this tutorial, you will learn
How to use DeepONet architecture in Modulus
How to set up data-informed and physics-informed DeepONet for learning operators
This tutorial assumes that you have completed the tutorial Introductory Example and are familiar with Modulus APIs.
Problem Description
Consider a 1D initial value problem
(176)\[\frac{du}{dx} = a(x), \quad x \in [0, 1],\]
subject to an initial condition \(u(0)=0\). The anti-derivative operator \(G\) over \([0,1]\) given by
(177)\[G:\quad a(x) \mapsto G(a)(x):= \int_0^x a(t) dt, \quad x \in [0,1].\]
You’re going to setup a DeepONet to learn the operator \(G\). In this case, the \(a\) will be the input of branch net and the \(x\) will be the input of trunk net. As the input of branch net, \(a\) is discretized on a fixed uniform grid. They are not necessary to be the same as the query coordinates \(x\) at which a DeepONet model is evaluated. For example, you may give the data of \(a\) as \(\{a(0),\ a(0.5),\ a(1)\}\) but evaluate the output at \(\{G(a)(0.1), G(u)(0.8), G(u)(0.9)\}\). This is one of the advantages of DeepONet compared with Fourier neural operator.
Data Preparation
As data preparation, generate \(10,000\) different input functions \(a\) from a zero mean Gaussian random field (GRF)
with an exponential quadratic kernel of a length scale \(l=0.2\). Then obtain the corresponding \(10,000\)
ODE solutions \(u\) using an explicit Runge-Kutta method. For each input output pair of \((a, u)\), it is worth noting
that only one observation of \(u(\cdot)\) is selected. It highlights the flexibility of DeepONet in terms
of tackling various data structure. The training and validation data are provided in /examples/anti_derivative/data/.
With this data, you can start the data informed DeepONet code.
The python script for this problem can be found at /examples/anti_derivative/data_informed.py.
Case Setup
Let us first import the necessary packages.
import torch
import numpy as np
import modulus
from modulus.hydra import to_absolute_path, instantiate_arch, ModulusConfig, to_yaml
from modulus.solver import Solver
from modulus.domain import Domain
from modulus.models.fully_connected import FullyConnectedArch
from modulus.models.fourier_net import FourierNetArch
from modulus.models.deeponet import DeepONetArch
from modulus.domain.constraint.continuous import DeepONetConstraint
from modulus.domain.validator.discrete import GridValidator
from modulus.dataset.discrete import DictGridDataset
from modulus.key import Key
Initializing the Model
In this case, you will use a fully-connected network as the branch net and a Fourier feature network as the trunk net.
In branch net, the Key("a", 100) and Key("branch", 128) specify the input and the output shape corresponding to one input function \(a\).
Similarly, in trunk net, the Key("x", 1) and Key("trunk", 128) specify the input and the output shape corresponding to one coordinate point \(x\).
In the config, these models are defined under the arch config group.
defaults:
- modulus_default
- /arch/fully_connected_cfg@arch.branch
- /arch/fourier_cfg@arch.trunk
- /arch/deeponet_cfg@arch.deeponet
- scheduler: tf_exponential_lr
- optimizer: adam
- loss: sum
- _self_
arch:
branch:
nr_layers: 4
layer_size: 128
trunk:
frequencies: "('axis',[iforiinrange(5)])"
nr_layers: 4
layer_size: 128
deeponet:
output_keys: u
scheduler:
decay_rate: 0.9
decay_steps: 100
training:
rec_validation_freq: 1000
max_steps: 10000
batch_size:
train: 10000
validation: 100
save_filetypes: "np"
The models are initialized in the Python script using the following:
trunk_net = instantiate_arch(
cfg=cfg.arch.trunk,
input_keys=[Key("x")],
output_keys=[Key("trunk", 128)],
)
branch_net = instantiate_arch(
cfg=cfg.arch.branch,
input_keys=[Key("a", 100)],
output_keys=[Key("branch", 128)],
)
deeponet = instantiate_arch(
cfg=cfg.arch.deeponet,
output_keys=[Key("u")],
branch_net=branch_net,
trunk_net=trunk_net,
)
nodes = [deeponet.make_node('deepo')]
The DeepONet architecture in Modulus is extremely flexible allowing users to use different branch and trunk nets. For example a convolutional model can be used in the branch network while a fully-connected is used in the trunk.
Loading Data
Then import the data from the .npy file.
# load training data
data = np.load(
to_absolute_path("data/anti_derivative.npy"), allow_pickle=True
).item()
x_train = data["x_train"]
a_train = data["a_train"]
u_train = data["u_train"]
# load test data
x_test = data["x_test"]
a_test = data["a_test"]
u_test = data["u_test"]
Adding Data Constraints
To add the data constraint, use DeepONetConstraint.
# make domain
domain = Domain()
data = DeepONetConstraint.from_numpy(
nodes=nodes,
invar={"a": a_train, "x": x_train},
outvar={"u": u_train},
batch_size=cfg.batch_size.train,
)
domain.add_constraint(data, "data")
Adding Data Validator
You can set validators to verify the results.
# add validators
for k in range(10):
invar_valid = {
"a": a_test[k * 100 : (k + 1) * 100],
"x": x_test[k * 100 : (k + 1) * 100],
}
outvar_valid = {"u": u_test[k * 100 : (k + 1) * 100]}
dataset = DictGridDataset(invar_valid, outvar_valid)
validator = GridValidator(nodes=nodes, dataset=dataset, plotter=None)
domain.add_validator(validator, "validator_{}".format(k))
Training the Model
Start the training by executing the python script.
python data_informed.py
Results
The validation results (ground truth, DeepONet prediction, and difference, respectively) are shown as below (Fig. 89, Fig. 90, Fig. 91).
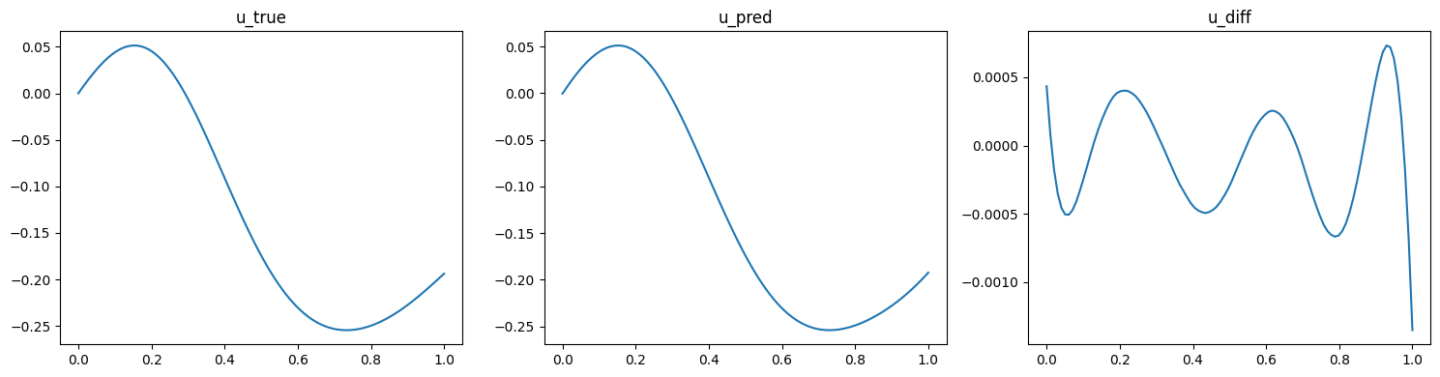
Fig. 89 Data informed DeepONet validation result, sample 1
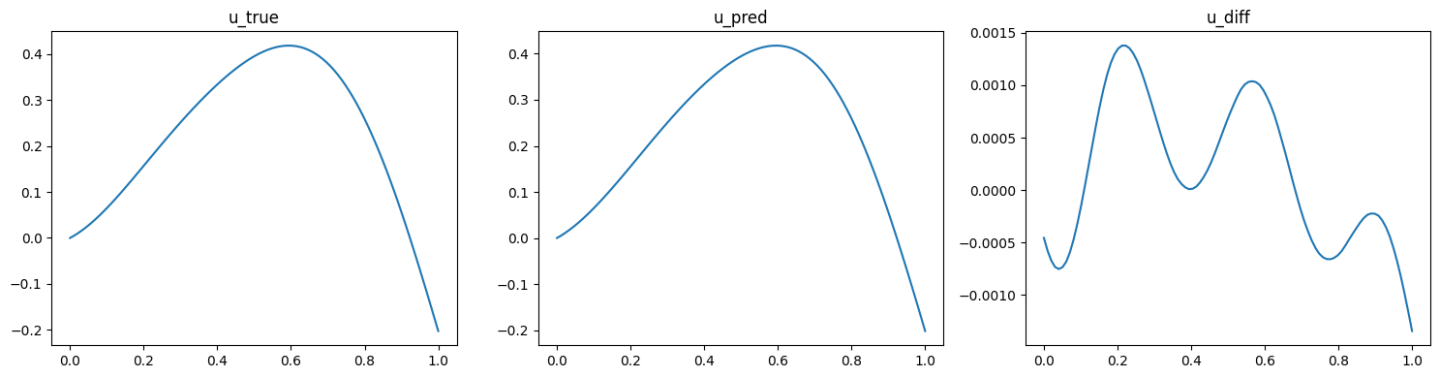
Fig. 90 Data informed DeepONet validation result, sample 2
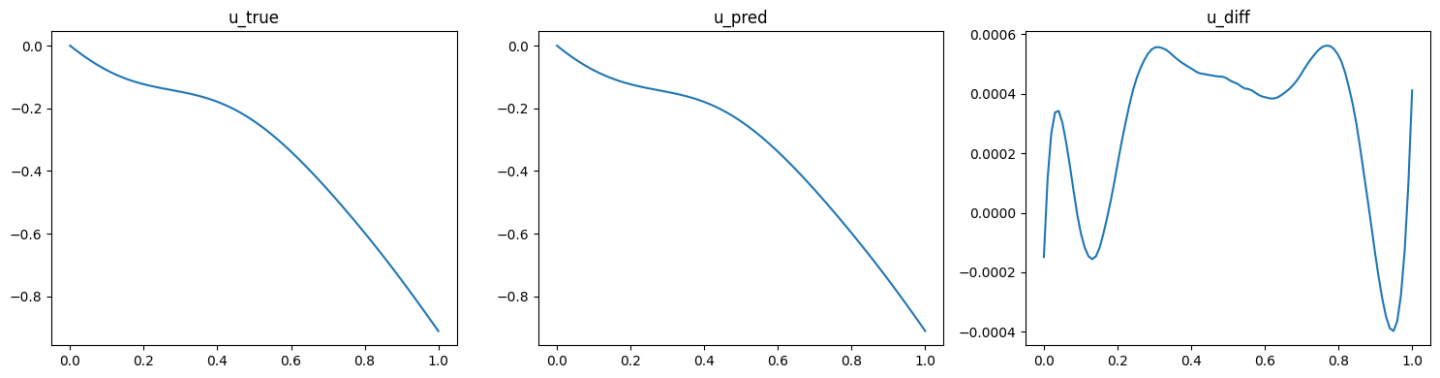
Fig. 91 Data informed DeepONet validation result, sample 3
This section uses the physics-informed DeepONet to learn the anti-derivative operator without any observations except for the given initial condition of the ODE system. Although there is no need for the training data, you will need some data for validation.
The python script for this problem can be found at /examples/anti_derivative/physics_informed.py.
Case Setup
Most of the setup for physics-informed DeepONet is the same as the data informed version. First you import the needed packages.
import torch
import numpy as np
import modulus
from modulus.hydra import to_absolute_path, instantiate_arch, ModulusConfig
from modulus.solver import Solver
from modulus.domain import Domain
from modulus.models.fully_connected import FullyConnectedArch
from modulus.models.fourier_net import FourierNetArch
from modulus.models.deeponet import DeepONetArch
from modulus.domain.constraint.continuous import DeepONetConstraint
from modulus.domain.validator.discrete import GridValidator
from modulus.dataset.discrete import DictGridDataset
from modulus.key import Key
Initializing the Model
In the run function, setup the branch and trunk nets, respectively. This part is the same as the data informed version.
# make list of nodes to unroll graph on
trunk_net = instantiate_arch(
cfg=cfg.arch.trunk,
input_keys=[Key("x")],
output_keys=[Key("trunk", 128)],
)
branch_net = instantiate_arch(
cfg=cfg.arch.branch,
input_keys=[Key("a", 100)],
output_keys=[Key("branch", 128)],
)
deeponet = instantiate_arch(
cfg=cfg.arch.deeponet,
output_keys=[Key("u")],
branch_net=branch_net,
trunk_net=trunk_net,
)
nodes = [deeponet.make_node('deepo')]
Loading Data
Then, import the data as the data informed version.
# load training data
data = np.load(
to_absolute_path("data/anti_derivative.npy"), allow_pickle=True
).item()
x_train = data["x_train"]
a_train = data["a_train"]
u_train = data["u_train"]
x_r_train = data["x_r_train"]
a_r_train = data["a_r_train"]
u_r_train = data["u_r_train"]
# load test data
x_test = data["x_test"]
a_test = data["a_test"]
u_test = data["u_test"]
Adding Constraints
Now the main difference of physics informed version compared with data informed is highlighted. First, impose the initial value constraint that \(a(0)=0\). The way to achieve this is to set the input of the trunk net as all zero data. Then the output function will be evaluated at only \(0\).
# make domain
domain = Domain()
# add constraints to domain
IC = DeepONetConstraint.from_numpy(
nodes=nodes,
invar={"a": a_train, "x": np.zeros_like(x_train)},
outvar={"u": np.zeros_like(u_train)},
batch_size=cfg.batch_size.train,
)
domain.add_constraint(IC, "IC")
Next, impose the derivative constraint that \(\frac{d}{dx}u(x) = a(x)\).
Note here that u__x is the derivative of u w.r.t x.
interior = DeepONetConstraint.from_numpy(
nodes=nodes,
invar={"a": a_r_train, "x": x_r_train},
outvar={"u__x": u_r_train},
batch_size=cfg.batch_size.train,
)
domain.add_constraint(interior, "Residual")
Adding Data Validator
Finally, add the validator. This is the same as data informed version.
# add validators
for k in range(10):
invar_valid = {
"a": a_test[k * 100 : (k + 1) * 100],
"x": x_test[k * 100 : (k + 1) * 100],
}
outvar_valid = {"u": u_test[k * 100 : (k + 1) * 100]}
dataset = DictGridDataset(invar_valid, outvar_valid)
validator = GridValidator(nodes=nodes, dataset=dataset, plotter=None)
domain.add_validator(validator, "validator_{}".format(k))
Training the Model
Start the training by executing the python script.
python physics_informed.py
Results
The validation results (ground truth, DeepONet prediction, and difference, respectively) are shown as below (Fig. 92, Fig. 93, Fig. 94).
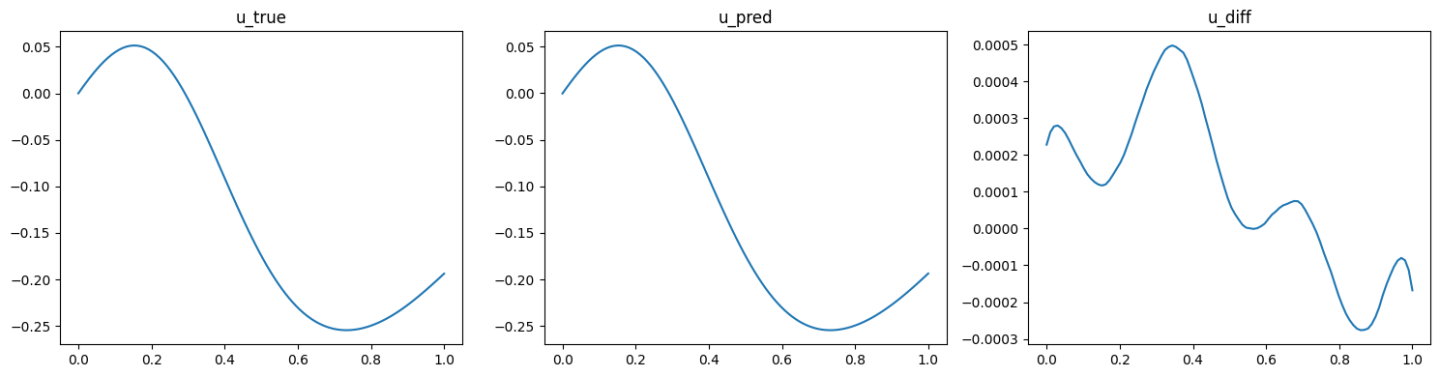
Fig. 92 Physics informed DeepONet validation result, sample 1
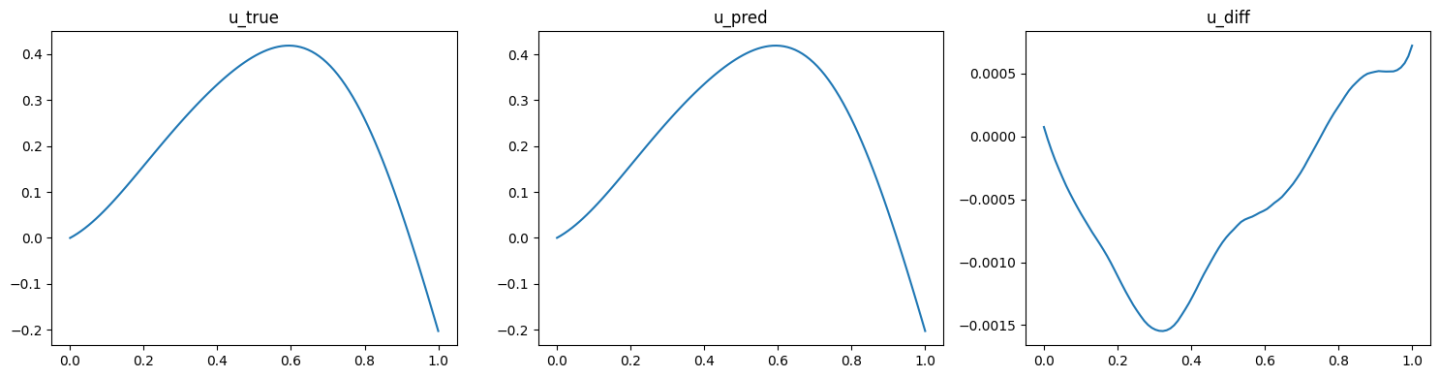
Fig. 93 Physics informed DeepONet validation result, sample 2
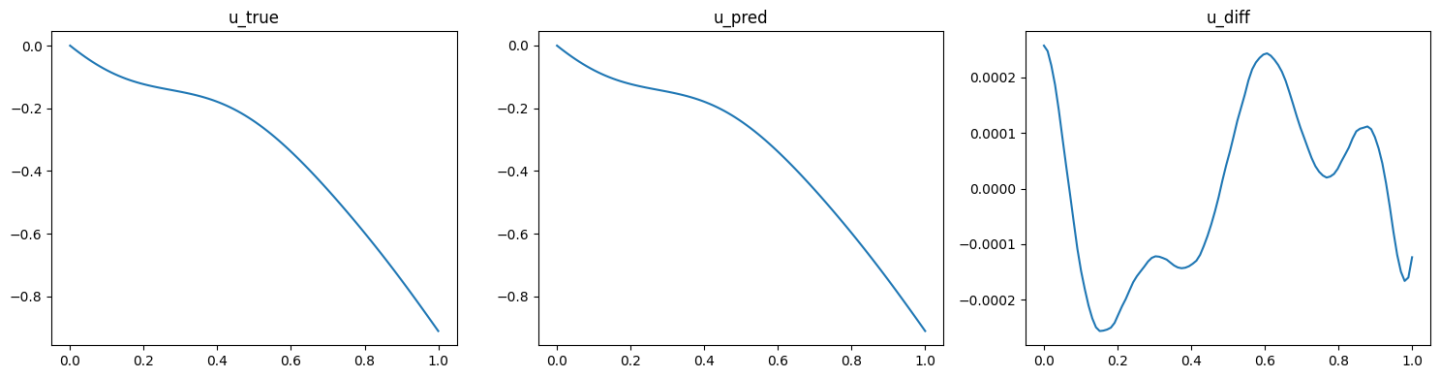
Fig. 94 Physics informed DeepONet validation result, sample 3
Case Setup
In this section, you will set up a data-informed DeepONet for learning the solution operator of a 2D Darcy flow in Modulus. The problem setup and training data are the same as in Fourier Neural Operators. Please see the tutorial Darcy Flow with Fourier Neural Operator for more details. It is worth emphasising that one can employ any built-in Modulus network architectures in a DeepONet model.
The python script for this problem can be found at examples/darcy/darcy_DeepO.py.
Loading Data
Loading both the training and validation datasets into memory follows a similar process as the Darcy Flow with Fourier Neural Operator example.
# load training/ test data
branch_input_keys = [Key("coeff")]
trunk_input_keys = [Key("x"), Key("y")]
output_keys = [Key("sol")]
download_FNO_dataset("Darcy_241", outdir="datasets/")
invar_train, outvar_train = load_deeponet_dataset(
"datasets/Darcy_241/piececonst_r241_N1024_smooth1.hdf5",
[k.name for k in branch_input_keys],
[k.name for k in output_keys],
n_examples=1000,
)
invar_test, outvar_test = load_deeponet_dataset(
"datasets/Darcy_241/piececonst_r241_N1024_smooth2.hdf5",
[k.name for k in branch_input_keys],
[k.name for k in output_keys],
n_examples=10,
)
Initializing the Model
Initializing DeepONet and domain is similar to the anti-derivative example but this time we will use a convolutional model. Similar to the FNO example the model can be configured entirely through the config file. A pix2pix convolutional model will be used as the branch net a while a fully-connected will be used as the trunk. The DeepONet architecture will automatically handle the dimensionality difference.
defaults:
- modulus_default
- /arch/pix2pix_cfg@arch.branch
- /arch/fourier_cfg@arch.trunk
- /arch/deeponet_cfg@arch.deeponet
- scheduler: tf_exponential_lr
- optimizer: adam
- loss: sum
- _self_
arch:
branch:
input_keys: [coeff]
output_keys: [branch]
dimension: 2
conv_layer_size: 32
trunk:
input_keys: [x, y]
output_keys: ['trunk', 256]
frequencies: "('axis',[iforiinrange(5)])"
nr_layers: 4
layer_size: 128
deeponet:
output_keys: sol
branch_dim: 1024
scheduler:
decay_rate: 0.9
decay_steps: 2000
training:
rec_validation_freq: 1000
max_steps: 100000
batch_size:
train: 1000
save_filetypes: "np"
The models are initialized inside the Python script using the following:
# make list of nodes to unroll graph on
branch_net = instantiate_arch(
cfg=cfg.arch.branch,
)
trunk_net = instantiate_arch(
cfg=cfg.arch.trunk,
)
deeponet = instantiate_arch(
cfg=cfg.arch.deeponet,
branch_net=branch_net,
trunk_net=trunk_net,
)
nodes = [deeponet.make_node(name="deepo")]
Adding Data Constraints and Validators
Then you can add data constraints as before
# make domain
domain = Domain()
# add constraint to domain
data = DeepONetConstraint.from_numpy(
nodes=nodes,
invar=invar_train,
outvar=outvar_train,
batch_size=cfg.batch_size.train,
)
domain.add_constraint(data, "data")
# add validators
val = PointwiseValidator(
nodes=nodes,
invar=invar_test,
true_outvar=outvar_test,
plotter=None,
)
domain.add_validator(val, "val")
Training the Model
The training can now be simply started by executing the python script.
python darcy_DeepO.py
Results and Post-processing
The validation results (ground truth, DeepONet prediction, and difference, respectively) are shown as below.
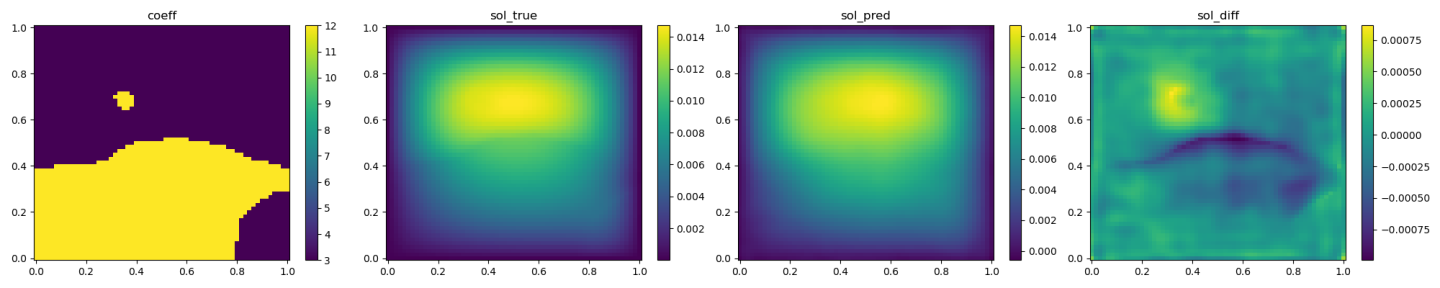
Fig. 95 DeepONet validation result, sample 1
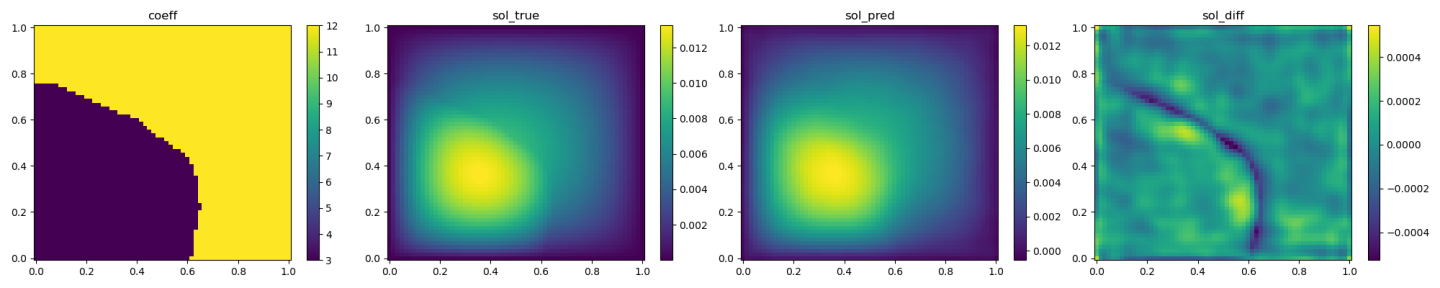
Fig. 96 DeepONet validation result, sample 2
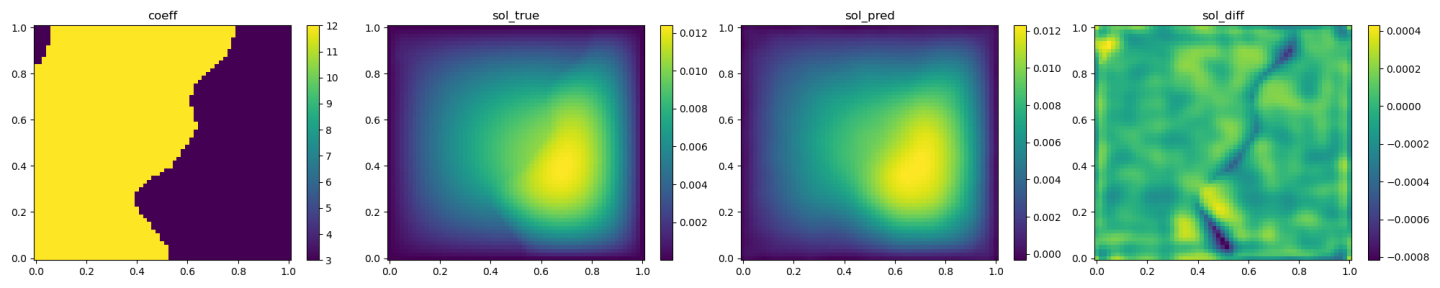
Fig. 97 DeepONet validation result, sample 3